Prevalence, Conventional and Molecular Characterization of Salmonella Isolated from Chicken Farms and Slaughterhouses
Research Article
Prevalence, Conventional and Molecular Characterization of Salmonella Isolated from Chicken Farms and Slaughterhouses
Mohammed H. Galhoum1, Hamza M. Eed2, Essam S. Soliman1*
1Abu Suwayr Veterinary Medical Unit, Directorate of Veterinary Medicine, Ismailia 41513, Egypt; 2Department of Bacteriology, Immunology, and Mycology, Faculty of Veterinary Medicine, Ain Shams University, Shubra Al Khaimah, Cairo 11241, Egypt; 3Department of Animal Hygiene, Zoonosis, and Animal Behavior, Faculty of Veterinary Medicine, Suez Canal University, Ismailia 41522, Egypt.
Abstract | The consumers’ perspective and expectations on food safety and quality are increased over time as many food stuff and products are exposed to contamination by numerous pathogens concerning Salmonella species. The current study aimed to investigate the prevalence of Salmonella contributing contamination in chicken samples that were collected from broiler farms and slaughterhouses using conventional culturing, biochemical, and serological identifications versus molecular detection. A prospective study was designed to last for six months from March 2021 to the end of August 2021. A total number of 126 chicken samples (100 samples from five broiler chicken farms and 26 samples from two slaughterhouses) were collected from the Ismailia governorate. Each sample was composed of liver, intestine, and breast and thigh muscles. The study revealed a total prevalence of 35.7% (45 positives out of 126 samples). Slaughterhouse I and II, chicken farms I, II, III, IV, and V revealed prevalence up to 15.3, 23.0, 40.0, 35.0, 50.0, 45.0, and 30% respectively. Initial isolation revealed 14.2% and delayed isolation procedures revealed a prevalence of up to 21.42%. The bacteriological analysis was carried out using conventional cultural and molecular means (cyclic polymerase chain reaction; cPCR) targeting the invA gene. The isolated Salmonella culture revealed higher resistance incidence up to 100% against amoxicillin-clavulanic acid (AMC; 30 μg), ampicillin (AMP; 10 μg), and nalidixic acid (NAL; 30 μg), 90% against enrofloxacin (ENR; 5 μg), and 80% against doxycycline HCL (DO; 30 μg). The conventional culture method revealed up to 83% sensitivity and 90% specificity while the molecular analysis revealed up to 100% sensitivity and 100% specificity for Salmonella detection. The study concluded that the high prevalence of the Salmonella with high resistance against 60% of the tested antibiotics reflects a serious problem with the hygienic and biosecurity measures taken in the poultry and slaughterhouses, as well the extensive use of the antibiotic contributed to the recorded high resistance among the isolated strains.
Keywords | Antimicrobial, Broiler chickens, Conventional, Molecular, Prevalence, Salmonella
Received | December 08, 2021; Accepted | December 21, 2021; Published | February 15, 2022
*Correspondence | Essam S Soliman, Department of Animal Hygiene, Zoonosis, and Animal Behavior, Faculty of Veterinary Medicine, Suez Canal University, Ismailia 41522, Egypt; Email: [email protected]
Citation | Galhoum MH, Eed HM, Soliman ES (2022). Prevalence, conventional and molecular characterization of Salmonella isolated from chicken farms and slaughterhouses. Adv. Anim. Vet. Sci. 10(3): 639-650.
DOI | http://dx.doi.org/10.17582/journal.aavs/2022/10.3.639.650
ISSN (Online) | 2307-8316
INTRODUCTION
The poultry industry directed its goals on combating infectious and contagious diseases, sustaining high production, raising the product quality, and achieving the goals with low costs (Cavani et al., 2009; Delpont et al., 2021; Schweitzer et al., 2021). To meet these expectations, good biosecurity measures have to be applied in the poultry facilities to minimize the entrance of pathogenic micro-organisms known as “bioexclusion” and prevent the transmission of the pathogens from one area to another known as “biocontainment” (Hafez, 2005; FAO Statitics, 2020; Delpont et al., 2021). Several actions have been adapted like increasing the self-sufficiency of broiler chickens, monitor and observing to increase the control over the disease’s development, increasing the veterinary services to improve the productivity and reduce the disease incidence, improving the vaccination act, and establishment of legislation programs to control the development of infectious diseases (Attia et al., 2017; Otte et al., 2021).
The consumers perspective on food safety and quality is a continuous issue. Many food stuff and products are exposed to contamination by numerous pathogens concerning Salmonella and Campylobacter species. The control of these pathogens should involve a deep understanding of the epidemiological triad of these micro-organisms and the commitment to the application of strict preventive and biosecurity measures (Alsultan et al., 2019; Morishita and Derksen, 2021). Many serious actions adapted and approved some actions to prevent the contamination of poultry products with pathogenic micro-organisms like Salmonella enterica serovars.
Salmonella is a genus of the family Enterobacteriaceae that includes more than 300 serovars and caused many disease problems in chickens (Sheela et al., 2003; Rogers et al., 2021). Salmonella is a gram-negative, non-spore-forming rod, motile through flagella but can shift into non-motile onto cultures (Su and Chiu, 2007; Pulford et al., 2021). Salmonella is chemotrophs that contain their energy from the oxidation-reduction reactions in the organic sources surrounding those (Rayan et al., 2017; Rosenberg et al., 2021). They are facultative anaerobes that are capable of generating adenosine triphosphate (ATP) from the oxygen once it can be available, otherwise, they use electron acceptors at the end of the transport chain including sulfate, nitrate, or sulfur, or fermentation (Fàbrega and Vila, 2013; Johansson et al., 2021). Salmonella is not heat resistant and at the same time doesn’t grow at low microclimatic temperatures, but they also may survive in an excellent state in acid foods and can as well resist dehydration. Meaning, while not able to multiply in many processed foods, if contamination is present, it can be difficult to eradicate (Mandal and Kwon, 2017).
The current study investigated the presence/absence information of Salmonella serovars in chicken samples collected from chicken farms and slaughterhouses. The investigation was based on conventional culturing means, biochemical, and serological identifications versus the molecular investigation using cyclic polymerase chain reaction (cPCR) targeting the invA gene.
MATERIALS AND METHODS
Ethical approval
The materials, methodology, and study design were approved by the Scientific Research Ethics Committee on animal and poultry researches, Faculty of Veterinary Medicine, Suez Canal University, Egypt with approval number (2021030).
The experimental design
A prospective study was designed to last for six months from March 2021 to the end of August 2021. The study was conducted to investigate the presence/absence information of Salmonella serovars in chicken meat collected from chicken farms at the marketing time, as well from slaughterhouses as marketable chickens passed for human consumption.
The samples were collected from five broiler chicken farms and two slaughterhouses located in the Ismailia governorate. Ismailia is situated on the west bank of the Suez Canal approximately halfway between the Port Said governorate to the north and Suez governorate to the south with a longitude of 30.5965° N and latitude of 32.2715° E. The climate in Ismailia according to the Köppen-Geiger climate classification system is known to be a hot desert. The hottest recorded temperature was 47°C (117°F) on 14 June while the coldest recorded temperature was 0.2°C (32.4°F) in January.
Sampling and sample preparations
A total number of 126 chicken samples were collected from the Ismailia governorate. The samples were collected at a rate of 100 samples from five broiler chicken farms and 26 samples from two different slaughterhouses. Each sample was composed of liver, intestine, and breast and thigh muscles. The samples were preserved to prevent any further contamination or decaying in an ice-box and transferred to the laboratory as quickly as possible where samples were kept frozen until bacteriological analysis.
In the laboratory, the samples were thawed carefully under complete aseptic conditions, and small pieces of the liver, duodenum of the intestine, and muscle tissues were dissected to be added to pre-enrichment tubes previously set containing 9 ml of buffered peptone water (Thermo Scientific™ Oxoid™ Buffered Peptone Water, CM0509B, 500 g) and incubated at 37°C for 18-24 hours as recommended by American Public Health Association; APHA (2017).
Bacteriological examination
Bacteriological examination was carried out following Herigstad et al. (2001) by transferring one ml from the pre-enriched samples under complete aseptic conditions into clean sterile tubes containing 9 ml fresh Rappaport Vassiliadis broth (RV, Thermo Scientific™ Oxoid™ Rappaport-Vassiliadis Enrichment Broth, CM0669, 500 g) and incubated at 37oC for 18-24 h. Ten µl were dropped onto already solidified Xylose Lysine Deoxycholate (XLD, Thermo Scientific™ Oxoid™ X.L.D. Agar, CM0469, 500 g) agar plates and incubated at 37oC for 18-24 h. The culturing was conducted using the drop plate technique as recommended by Soliman et al. (2016) and Kim and Lee (2016). The plates were examined morphologically for the growth of the typical black colonies as recommended by Murray et al. (2015).
Negative samples were subjected to delaying protocol through additional enrichment of one ml from buffered peptone water tubes into 9 ml tubes of Rappaport Vassiliadis broth at room temperature for 5-7 days in closed sterile colorless glass bottles with daily renewal of the Rappaport Vassiliadis broth to prevent the desiccation and decaying of the samples. Later, the delayed samples were processed by transferring 1 ml under complete aseptic conditions into clean sterile tubes containing fresh RV broth and incubated at 37°C for 18-24 h. Ten µl were dropped onto already XLD agar plates and incubated at 37°C for18-24 h. The plates were examined for the development of black colonies. The colonies were streaked for biochemical identification, antimicrobial sensitivity testing, and some colonies were preserved into RV broth with glycerol into sterile 2.5 ml Eppendorf tubes for serological identification and RV broth into sterile serum tubes for polymerase chain reaction identification.
Biochemical identification
The biochemical identification was carried out using serious biochemical tests like the triple sugar iron agar test (TSI) indicating gas production and changes in the color from red to yellow. Lysine iron agar test (LIA) to determine the ability of the micro-organism to deaminate lysine aerobically on the slant of the media or anaerobically decarboxylate lysine in the butt of the media.
The urease test to determine the microbial capabilities of hydrolyzing urea to produce ammonia and carbon dioxide. The indole production test was used to measure the ability of micro-organisms to decompose the amino acid tryptophan to indole which accumulates in the medium. Methyl red test (MR) was used to determine the microbial abilities for the production of acid as it identifies bacterial ability to produce stable acid end products through a mixed-acid fermentation of glucose. Voges Proskauer test (VP) determined if an organism produces acetyl methyl carbinol from glucose fermentation.
Serological identification
The isolated Salmonella isolates were serotyped using slide agglutination test (stained Salmonella antigen Widal latex slide test kit, 8 × 5 mL, Bio Lab® Diagnostics (I) Private Limited) according to Collins et al. (1995). The suspected colonies were sub-cultured on nutrient slopes at 37°C for 24 hours. A slide agglutination test was carried out by re-suspending Salmonella colonies into two separate drops of sterile physiological saline on a slide. A drop of Salmonella somatic antigen “Salmonella O” and Salmonella flagellar antigen “Salmonella H” were added to the suspended colonies with thorough mixing. Positive results can be detected by the development of agglutinations that can be seen by the naked eyes within a minute. The delayed agglutinations or homogenous drops indicated negative results.
Molecular identification
Extraction of DNA (QIAamp DNA Mini Kit, Catalogue no. 51304)
A mix of 20 μl QIAGEN protease, 200 μl of the sample, and 200 μl buffer AL were pipetted into the bottom of a 1.5 ml micro-centrifuge tubes, vortex for 15 sec, and incubated at 56˚C for 10 min. About 200 μl of ethanol (96%) were added and mixed by pulse vortex for 15 sec. Then mixtures were carefully transferred to the QIAamp Mini spin column in a 2ml collecting tube and centrifuged at 8000 rpm for one min. The tubes containing the filtrate were discarded and 500 ml from buffer AW1 were added, centrifuged (8000 rpm/ one min), 500 ml buffer AW2 were added and centrifuged at full speed for 3 min, and a 100 μl from buffer AE were added, incubated at room temperature (15-25˚C) for one min, and then centrifuged at 8000 rpm for one min.
Preparation of the master-mix
The master-mix was prepared according to Emerald Amp GT PCR master-mix (Takara®) Code No. RR310A kit. The tubes were set with Emerald Amp GT PCR master-mix (2x premix); 12.50 μl, PCR grade water; 4.50 μl, forward primer (20 pmol); 1.00 μl, reverse primer (20 pmol); 1.00 μl, template DNA; 6.00 μl, and the total reaction was optimized at 25.00 μl.
Oligonucleotide primer sequences
The primers were designed (Metabion®, Germany) as follows:
5′-GTGAAATTATCGCCACGTTCGGGCAA-3′.
3′-TCATCGCACCGTCAAAGGAACC-5′.
The primers were targeting the invA gene of Salmonella (Oliveira et al., 2003). The produced amplified product was 284 bp.
Cycling conditions of the primers during cPCR
The temperature and time conditions of the two primers for detecting the invA gene of Salmonella during the PCR were as follow: primary denaturation at 94°C/5 min, secondary denaturation at 94°C/30 sec, annealing at 55°C/30 sec, extension at 72°C/30 sec, the total number of cycles was designed to 35 cycles (secondary denaturation, annealing, and extension), and the final extension at 72°C/7 min.
Agarose gel electrophoresis
The gel electrophoresis was carried out according to Sambrook et al. (1989). Ten μl of the required ladder were directly loaded. Electrophoresis grade agarose (1.0 g) was prepared in 100 ml TBE buffer, heated in a microwave to dissolve, allowed to cool at 70˚C, then 0.5 μg/ml Ethidium bromide was added, and mixed thoroughly. The warm agarose was poured directly into the gel casting apparatus with the desired comb in apposition and left at room temperature for polymerization.
The comb was then removed and the electrophoresis tank was filled with TBE buffer. Twenty μl of each uniplex PCR product, negative control, and positive control were loaded to the gel. The power supply was run at 1-5 volts/cm of the tank length. The run was stopped after about 30 min and the gel was transferred to the UV cabinet. The gel was photographed by a gel documentation system and the data was analyzed through computer software.
Antibiotic sensitivity
The antibiotic sensitivity test was carried out according to CLSI (Clinical and Laboratory Standards Institute, 2002). A single colony of the suspected colonies of Salmonella was inoculated into 5 ml tryptic soy broth and incubated at 37C for 18 hours. The turbidity of the tube was measured against 0.5 McFarland of 1.5 x 105 CFU/ml. Few drops of the turbid broth were inoculated onto Muller-Hinton agar plates. Excess of cultural fluid was removed aseptically and the plates were allowed to stand for at 37°C for 15 min for dryness. The inoculated plates were overlaid with antibiotic discs (Amoxicillin and Clavulanic acid - AMC; 30 μg, Ampicillin - AMP; 10 μg; Amikacin - AK; 30 μg; Doxycycline HCL - DO; 30 μg, Meropenem - MEM; 10 μg, Gentamicin - GN; 10 μg, Norfloxacin - NOR; 10 μg; Trimethoprim-sulfamethoxazole - SXT; 25 μg, Nalidixic acid - NAL; 30 μg, and Enrofloxacin - ENR; 5 μg) using sterile forceps considering the distribution of the discs in a manner where the distance among them was optimum and away from the edge of the plate to avoid overlapping of the inhibition zones and gives more wide area for the zone of inhibition.
The inoculated plates were incubated at 37°C for 24 hours. Inhibition zones were measured by caliper and interpretation of the results was carried out in comparison to the interpretative standards of the National Committee for Clinical Laboratory Standards (NCCLS, 1990, MZ-A4).
Sensitivity and specificity
The sensitivity referred to the proportion of those who have the condition that received a positive result on this test (Proportion of true positive). Sensitivity (Sn) was measured as recommended by Powers (2011) and Bénard et al. (2018) according to the following formula:
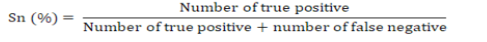
The specificity referred to the proportion of those do not who have the condition that received a negative result on this test (proportion of true negative). Specificity (Sp) was measured as recommended by Powers (2011) and Bénard et al. (2018) according to the following formula:
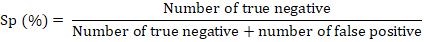
Statistical analysis
The statistical analysis was conducted using a statistical package for social sciences version 20 (IBM Corp, 2016 - IBM SPSS Statistics 20). The obtained data and results were analyzed statistically using One-Way Analysis of Variance (ANOVA) to estimate the prevalence and their statistical differences. The statistical model empathized as follow:
Yij = µ + αj + Ɛij
Where Yij was the measurement of dependent variables; µ was the overall mean; αj was the fixed effect of the bacteriological detection of samples positivity, and Ɛij was the random error. Nonparametric Kruskal–Wallis was used for detecting the significant differences between the prevalence rates. The results were expressed as highly significant at (p ≤ 0.01), significant at (p ≤ 0.05), and non-significant at (p > 0.05).
RESULTS and Discussion
Prevalence of Salmonella in chicken samples
The results in Table 1 revealed a total prevalence of 35.7% (45 positives out of 126 samples). This prevalence was variable among the sources from which samples have been collected. The different locations of sampling; slaughterhouse I and II, chicken farms, I, II, III, IV, and V revealed prevalence up to 15.3 (2 positives out of 13 samples), 23.0 (3 positives out of 13 samples), 40.0 (8 positives out of 20 samples), 35.0 (7 positives out of 20 samples), 50.0 (10 positives out of 20 samples), 45.0 (9 positives out of 20 samples), and 30% (6 positives out of 20 samples). The tissue-specific total prevalence revealed in Table 1 high isolation rates of Salmonella from the intestine (17.46%), muscles (11.90%), and liver (6.35%) samples, respectively. The higher isolation rates were detected in chicken farms’ samples compared to slaughterhouses samples.
Initial isolation procedures (Table 2) revealed 18 positive out of 126 samples with a prevalence of up to 14.2%. The prevalence of isolations was nearly zero (0%) in slaughterhouses’ samples compared to 15, 20, 25, 15, and 15% isolation rates from the five chicken farms (I, II, III, IV, and V), respectively. The tissue-specific initial culturing prevalence revealed in Table 2 high isolation rates of Salmonella from muscles (6.35%), intestine (6.35%), and liver (3.17%) samples, respectively.
Table 1: Prevalence of total and tissue-specific Salmonella positive samples during the study.
| Source | Tissues | No. | Positive | P (%) |
| Slaughterhouse I | Liver | 13 | 0 |
0.00b |
| Intestine | 13 | 1 |
7.69a |
|
| Muscles | 13 | 1 |
7.69a |
|
| Total | 13 | 2 |
15.38D |
|
| Slaughterhouse II | Liver | 13 | 0 |
0.00c |
| Intestine | 13 | 2 |
15.38a |
|
| Muscles | 13 | 1 |
7.69b |
|
| Total | 13 | 3 |
23.07D |
|
| Chicken farm I | Liver | 20 | 1 |
5.00c |
| Intestine | 20 | 4 |
20.00a |
|
| Muscles | 20 | 3 |
15.00b |
|
| Total | 20 | 8 |
40.00B |
|
| Chicken farm II | Liver | 20 | 1 |
5.00b |
| Intestine | 20 | 3 |
15.00a |
|
| Muscles | 20 | 3 |
15.00a |
|
| Total | 20 | 7 |
35.00C |
|
| Chicken farm III | Liver | 20 | 2 |
10.00c |
| Intestine | 20 | 5 |
25.00a |
|
| Muscles | 20 | 3 |
15.00b |
|
| Total | 20 | 10 |
50.00A |
|
| Chicken farm IV | Liver | 20 | 3 |
15.00b |
| Intestine | 20 | 4 |
20.00a |
|
| Muscles | 20 | 2 |
10.00c |
|
| Total | 20 | 9 |
45.00B |
|
| Chicken farm V | Liver | 20 | 1 |
5.00c |
| Intestine | 20 | 3 |
15.00a |
|
| Muscles | 20 | 2 |
10.00b |
|
| Total | 20 | 6 |
30.00C |
|
| Total | Liver | 126 | 8 |
6.35c |
| Intestine | 126 | 22 |
17.46a |
|
| Muscles | 126 | 15 |
11.90b |
|
| Total | 126 | 45 | 35.71 |
a,b,c,d,e Means carrying different superscripts in the same column are significantly different at (P ≤ 0.05) or highly significantly different at (P < 0.01). Means carrying the same superscripts in the same column are non-significantly different at (P < 0.05). A, B, C, D, E Means carrying different superscripts in the same column are significantly different at (P ≤ 0.05) or highly significantly different at (P < 0.01). Means carrying the same superscripts in the same column are non-significantly different at (P < 0.05). P= Prevalence (%).
Table 2: Prevalence of total and tissue-specific Salmonella positive samples in initial culturing.
| Source | Tissues | No. | Positive | P (%) |
| Slaughterhouse I | Liver | 13 | 0 |
0.00a |
| Intestine | 13 | 0 |
0.00a |
|
| Muscles | 13 | 0 |
0.00a |
|
| Total | 13 | 0 |
0.00D |
|
| Slaughterhouse II | Liver | 13 | 0 |
0.00a |
| Intestine | 13 | 0 |
0.00a |
|
| Muscles | 13 | 0 |
0.00a |
|
| Total | 13 | 0 |
0.00D |
|
| Chicken farm I | Liver | 20 | 0 |
0.00c |
| Intestine | 20 | 1 |
5.00b |
|
| Muscles | 20 | 2 |
10.00a |
|
| Total | 20 | 3 |
15.00C |
|
| Chicken farm II | Liver | 20 | 1 |
5.00b |
| Intestine | 20 | 1 |
5.00b |
|
| Muscles | 20 | 2 |
10.00a |
|
| Total | 20 | 4 |
20.00B |
|
| Chicken farm III | Liver | 20 | 1 |
5.00b |
| Intestine | 20 | 2 |
10.00a |
|
| Muscles | 20 | 2 |
10.00a |
|
| Total | 20 | 5 |
25.00A |
|
| Chicken farm IV | Liver | 20 | 1 |
5.00a |
| Intestine | 20 | 1 |
5.00a |
|
| Muscles | 20 | 1 |
5.00a |
|
| Total | 20 | 3 |
15.00C |
|
| Chicken farm V | Liver | 20 | 1 |
5.00a |
| Intestine | 20 | 1 |
5.00a |
|
| Muscles | 20 | 1 |
5.00a |
|
| Total | 20 | 3 |
15.00C |
|
| Total | Liver | 126 | 4 |
3.17b |
| Intestine | 126 | 6 |
4.76b |
|
| Muscles | 126 | 8 |
6.35a |
|
| Total | 126 | 18 | 14.28 |
a,b,c,d,e Means carrying different superscripts in the same column are significantly different at (P ≤ 0.05) or highly significantly different at (P < 0.01). Means carrying the same superscripts in the same column are non-significantly different at (P < 0.05). A, B, C, D, E Means carrying different superscripts in the same column are significantly different at (P ≤ 0.05) or highly significantly different at (P < 0.01). Means carrying the same superscripts in the same column are non-significantly different at (P < 0.05). Initial culturing included pre-enrichment on RV broth/ day and culturing on XLD agar/ day. P= Prevalence (%)
Delayed isolation procedures in Table 3 revealed a total of 27 positives out of 126 samples with a prevalence up to 21.42%. The isolation rates and prevalence were fluctuating with a minimum of 15% in chicken farm II to a maximum of 30% in chicken farm IV. The tissue-specific delayed culturing prevalence revealed in Table 3 high isolation rates of Salmonella from the intestine (12.70%), muscles (5.55%), and liver (3.17%) samples, chronologically.
Table 3: Prevalence of total and tissue-specific Salmonella positive samples in delayed culturing.
| Source | Tissues | No. | Positive | P (%) |
| Slaughterhouse I | Liver | 13 | 0 |
0.00b |
| Intestine | 13 | 1 |
7.69a |
|
| Muscles | 13 | 1 |
7.69a |
|
| Total | 13 | 2 |
15.38C |
|
| Slaughterhouse II | Liver | 13 | 0 |
0.00c |
| Intestine | 13 | 2 |
15.38a |
|
| Muscles | 13 | 1 |
7.69b |
|
| Total | 13 | 3 |
23.07B |
|
| Chicken farm I | Liver | 20 | 1 |
5.00b |
| Intestine | 20 | 3 |
15.00a |
|
| Muscles | 20 | 1 |
5.00b |
|
| Total | 20 | 5 |
25.00B |
|
| Chicken farm II | Liver | 20 | 0 |
0.00c |
| Intestine | 20 | 2 |
10.00a |
|
| Muscles | 20 | 1 |
5.00b |
|
| Total | 20 | 3 |
15.00C |
|
| Chicken farm III | Liver | 20 | 1 |
5.00b |
| Intestine | 20 | 3 |
15.00a |
|
| Muscles | 20 | 1 |
5.00b |
|
| Total | 20 | 5 |
25.00B |
|
| Chicken farm IV | Liver | 20 | 2 |
10.00b |
| Intestine | 20 | 3 |
15.00a |
|
| Muscles | 20 | 1 |
5.00c |
|
| Total | 20 | 6 |
30.00A |
|
| Chicken farm V | Liver | 20 | 0 |
0.00c |
| Intestine | 20 | 2 |
10.00a |
|
| Muscles | 20 | 1 |
5.00b |
|
| Total | 20 | 3 |
15.00C |
|
| Total | Liver | 126 | 4 |
3.17b |
| Intestine | 126 | 16 |
12.70a |
|
| Muscles | 126 | 7 |
5.55b |
|
| Total | 126 | 27 | 21.42 |
a,b,c,d,e Means carrying different superscripts in the same column are significantly different at (P ≤ 0.05) or highly significantly different at (P < 0.01). Means carrying the same superscripts in the same column are non-significantly different at (P < 0.05). A, B, C, D, E Means carrying different superscripts in the same column are significantly different at (P ≤ 0.05) or highly significantly different at (P < 0.01). Means carrying the same superscripts in the same column are non-significantly different at (P < 0.05). Delayed culturing included pre-enrichment on RV broth / five to seven days and culturing on XLD agar/ day. P= Prevalence (%).
Biochemical identifications
The biochemical profile revealed in Figure 1A positive yellow TSI slants (yellow coloration from the acid formation interrupted with gases and black color from the hydrogen sulfide formation), Figure 1B positive LIA slants (purple slants and purple but accompanied with blackening of the butt by hydrogen sulfide production), Figure 1C positive SC utilization test (blue color), and Figure 1D positive MR test (red color development). Meanwhile, Salmonella isolates were negative to all of the urease, indole production, and VP tests.
Serological identification
The serological identification using agglutination technique was carried out to all the positive samples from the traditional culturing technique (35.7%; 45 positives out of 126 samples) and revealed a majority of Salmonella typhi O by 84% and Salmonella typhi H by 16%.
Antibiotic sensitivity
The antibiotic sensitivity diffusion test against the isolated Salmonella culture revealed in Table 4 higher sensitivity incidence up to 100% to Amikacin (AK, 30 μg), Meropenem (MEM, 10 μg), and Gentamicin (GN, 10 μg) followed by lower sensitivity incidence up to 80% in Norfloxacin (NOR, 10 μg) and Trimethoprim-sulfamethoxazole (SXT, 25 μg). The isolated Salmonella culture revealed in Table 4 higher resistance incidence up to 100% against Amoxicillin- Clavulanic acid (AMC, 30 μg), Ampicillin (AMP, 10 μg), and Nalidixic acid (NAL, 30 μg) followed by a resistance level up to 90% against Enrofloxacin (ENR, 5 μg) and a resistance level up to 80% against Doxycycline HCL (DO, 30 μg).
Table 4: Incidence of antibacterial resistance in the isolated Salmonella.
| Antibiotic discs | Sensitivity |
| Amoxicillin- Clavulanic acid (AMC, 30 μg) | R (100%) |
| Ampicillin (AMP, 10 μg) | R (100%) |
| Amikacin (AK, 30 μg) | S (100%) |
| Doxycycline HCL (DO, 30 μg) | R (80%) |
| Meropenem (MEM, 10 μg) | S (100%) |
| Gentamicin (GN, 10 μg) | S (100%) |
| Norfloxacin (NOR, 10 μg) | S (80%) |
| Trimethoprim-sulfamethoxazole (SXT, 25 μg) | S (80%) |
| Nalidixic acid (NAL, 30 μg) | R (100%) |
| Enrofloxacin (ENR, 5 μg) | R (90%) |
Molecular identification
The molecular analysis was carried out on a representative sample size from the total collected samples (including the positive and negative samples from the culturing technique). The gel electrophoresis as shown in Figure 2 revealed about 80% positivity from the samples used in the traditional cultures. The positive bands were revealed at 284 bp compared to the ladder, positive, and negative control.
Sensitivity and specificity
The sensitivity (%) calculations revealed in Table 5 a total of 83% sensitivity (9 false positives) for the traditional culture method and 100% sensitivity (zero false positives) for the molecular detection of the Salmonella in the admitted samples. The specificity (%) revealed in Table 5 a total of 90% specificity (9 false negatives) for the traditional culture method and 100% specificity (zero false negatives) for the molecular detection of the Salmonella in the admitted samples.
Table 5: Sensitivity (Sn) and specificity (Sp) of the characterization methods.
| Items | Traditional cultures | PCR |
| Total no. of samples | 126 | 126 |
| No. of positives | 45 | 38 |
| False positives | 9 | zero |
| No. of negatives | 81 | 88 |
| False negatives | 9 | zero |
| Sensitivity (Sn) | 83% | 100% |
| Specificity (Sp) | 90% | 100% |
DISCUSSION
Salmonella is the most dangerous and worldwide foodborne pathogen and contributed to salmonellosis in animals, poultry, and human due to its zoonotic potential (Antunes et al., 2016; Center for Disease Control and Prevention, 2020). Salmonella usually can achieve access to humans via consumption of contaminated food causing non-typhoidal salmonellosis with severe gastro-enteritis manifestation (Braden, 2006). Salmonella usually gains access to contaminate poultry meat and products from the initial Salmonella infection in poultry farms from feed, water, wild birds, recovering birds, asymptomatic birds, rodents, and flies (Le Bouquin et al., 2010).
The current study showed a total prevalence of isolated Salmonella up to 35.7%. The different sampling locations like slaughterhouse I and II, chicken farms, I, II, III, IV, and V revealed prevalence up to 15.3, 23.0, 40.0, 35.0, 50.0, 45.0, and 30% respectively. The isolated Salmonella was detected by initial isolation procedures (18 positives out of 126 samples; 14.2%) and delayed isolation procedures (27 positives out of 126 samples; 21.42%). The results were compatible with those recorded by Donado-Godoy et al. (2012) who recorded a prevalence of up to 41% of Salmonella in broiler farms compared to the 40.5% median prevalence of Salmonella estimated worldwide. Rodriguez et al. (2015) and Kloska et al. (2017) also recorded a 17.4% prevalence of Salmonella in broiler production lines. They also strengthen that the authorities must consider the surveys on Salmonella status in the farms and ensure the appropriate biosecurity and control measures to be taken in poultry farms. Althaus et al. (2017) and Cota et al. (2019) showed that the poultry meat contamination might arise from the handling during slaughtering or using contaminated equipment and benches. The outcome ensures poor hygienic conditions and bad biosecurity measures.
Elshebrawy et al. (2021) also determined the prevalence of Salmonella serovars isolated from duck, pigeon, and quail carcasses in Egypt. They detected up to 62%, 40%, and 46% Salmonella in duck, pigeon, and quail carcasses, respectively with an overall prevalence of 49.3% (148/300). The current results also considered the different contamination sources that might arise from poor bio-security practices, depopulation time, litter recycling, inefficient flaming process, dust material (dust-borne infection), the hatchery machines, contaminated parental flocks, flock to flock infection-transmission, other biological reservoirs (pets or pests), and bad sanitation and disinfection practices as documented by Armwood et al. (2019), McWhorter et al. (2019), and Voss-Rech et al. (2019). Mir et al. (2010) examined 480 samples from adult health birds and tissue samples from four governmental farms in Kashmir from September 2007 to April 2008. They recovered Salmonella Gallinarum (84.8%), Salmonella Enteritidis (9.09%), and Salmonella Typhimurium (6.06%).
The serological identification on the positive colonies (35.7%; 45 positives out of 126 samples) revealed a majority of Salmonella typhi O by 84% and Salmonella typhi H by 16%. The results were in agreement with Rodriguez et al. (2015) who recorded 17.4% Salmonella that was serotyped as Salmonella paratyphi B with a rate up to 36.17% from the isolated serotypes in broiler production lines. Schneid et al. (2006) also evaluated the usage of an indirect enzyme-linked immune-sorbent assay (ELISA) based on a monoclonal antibody specific for Salmonella enterica serovar Enteritidis on 154 chicken meat samples. They revealed that 23% of the samples were contaminated according to the conventional cultures and 26% according to ELISA. They revealed higher sensitivity and specificity (94%) of ELISA compared to the conventional means.
The isolated Salmonella culture revealed higher resistance up to 100% against AMC (30 μg), AMP (10 μg), and NAL (30 μg) followed by a resistance level up to 90% against ENR (5 μg), and a resistance level up to 80% against DO (30 μg). The results were synchronized with those reported by Matsui et al. (2021) who recorded an increase in the proportion of Samonella Schwarzengrund resistant to kanamycin by a rate of 51.4–89.7%. Uddin et al. (2021) elucidate the molecular mechanisms, genetic relationships, and phenotype correlations of colistin-resistant Salmonella and found that the majority of the tested Salmonella isolates were found resistant to colistin (92.68%), ciprofloxacin (73.17%), tigecycline (62.20%) and trimethoprim/sulfamethoxazole (60.98%). Al-Ansaria et al. (2021) also reported that Salmonella zoonosis and infection presented hazardous effects on consumers and also recorded that the isolated serovars contributed to a high genotypic resistance pattern to antibiotics. That is why they recommended the restrictions of using antibiotics in poultry farms.
Obe et al. (2021) determined the antimicrobial tolerance of 25 Salmonella isolates recovered from poultry handling equipment and recorded minimum inhibitory concentration values between 500 and 1,000 parts per million for chlorine or 3 to 25 parts per million for quaternary ammonium compounds. Besides, the recorded isolates were resistant to multiple antibiotics, and 64% exhibited resistance to aminoglycosides and β-lactams. Yu et al. (2021) recorded high antimicrobial resistance of Salmonella enterica subsp. enterica serotype Enteritidis against nalidixic acid (97.6%) and ampicillin (74.2%) and they contributed this resistance to the genome structure of the Salmonella that arboured single mutations in gyrA, possessed the plasmid-mediated quinolone resistance genes qnrS (0.8%), oqxAB (2.4%), and the blaTEM-1 (67.7%), as well the extended-spectrum beta-lactamase (ESBL) genes blaCTX-M-55 were detected in 2.4% of the strains.
Hyeon et al. (2011) recorded that 18 Salmonella strains with Salmonella London and Salmonella Montevideo predominating in chicken meat revealed high resistance to erythromycin (100%), streptomycin (22.2%), tetracycline, and chloramphenicol (16.7%). Yildirim et al. (2011) detected Salmonella in 34% of the examined 200 packaged fresh raw chicken samples between April 2005 and March 2006 and identified ten Salmonella serovars including predominating Salmonella Typhimurium, Infantis, and Heidelberg. They also recorded high resistance against penicillin (100%), oxacillin (97%), clindamycin (97%), vancomycin (92.6%), erythromycin (89.7%), ampicillin (85.2%), tetracycline (67.6%), streptomycin (61.7%), neomycin (55.8%), and cephalothin (52.9%). They concluded that strict hygienic practices have to be enforced to reduce the high contamination levels. Thai and Yamaguchi (2012) examined 283 samples from retail meat and isolated 118 Salmonella isolates including Infantis, Anatum, Rissen, Reading, Emek, Typhimurium, Blockley, London, Newport, Derby, Weltevreden, Albany, and Hadar. They revealed tetracycline (54.2%), sulfonamides (52.5%), streptomycin (41.5%), trimethoprim (36.4%), chloramphenicol (35.6%), and ampicillin (33.1%).
The current results revealed the positive bands in the gel electrophoresis of the molecular outcome at 284 bp compared to the ladder, positive, and negative control. The traditional culture method revealed up to 83% sensitivity and 90% specificity while the molecular analysis (targeting the invA gene) revealed up to 100% sensitivity and 100% specificity for Salmonella detection in the admitted samples. The results were in agreement with those reported by Rodríguez-Hernández et al. (2021) who collected a total of 135 samples from 15 broiler farms (cloacal, feed, water, environmental, and farm operator feces samples) and they carried molecular confirmation of Salmonella isolates by amplification of the invA gene and they identified the isolates as Salmonella paratyphi B. Gand et al. (2019) also validated the molecular confirmation and characterization for 178 Salmonella paratyphi B with accuracy up to 100% compared to biochemical testing and 98% compared to the serological identification.
Davanzo et al. (2021) confirmed molecular characterization as a highly sensitive technique for the detection and characterization of Salmonella in poultry slaughterhouses. Kagambega et al. (2021) recorded the efficiency of the used technique for determining 111 strains of Salmonella isolated from poultry feces in Burkina Faso using a multiplex assay for rapid typing (SMART) polymerase chain reaction (PCR).
Hyeon et al. (2011) identified 18 Salmonella strains as Salmonella London and Salmonella Montevideo predominating in chicken meat in a study on the prevalence of Salmonella in chicken, beef, and pork meat from wholesale markets, retail stores, and traditional markets in Seoul, South Korea, in 2009 using rep-PCR except in Salmonella London and Montevideo. Abd El-Aziz (2013) examined 100 retail raw meat and giblets samples in Assiut governorate-Egypt and recorded Salmonella Typhimurium at a rate of 44%, 40%, and 48% in chicken meat, liver, and heart, respectively using duplex PCR amplification of DNA using rfbJ and fliC genes. Schneid et al. (2006) recorded nearly similar results when compared to the traditional cultures to the serological detection of Salmonella enterica serovar Enteritidis on 154 chicken meat samples, they revealed higher sensitivity and specificity (94%) of ELISA compared to the conventional means.
CONCLUSION and RECOMMENDATIONS
A high prevalence of up to 35.7% (14.2% from the initial enrichment and 21.5% from the delayed procedures) of the isolated Salmonella was detected in the study period among the five broiler farms and the two slaughterhouses. The isolated Salmonella revealed high resistance up to 80: 100% against many of the tested antibiotics (AMC 30 μg, AMP 10 μg, and NAL 30 μg, ENR 5 μg, DO 30 μg). The conventional culture method revealed up to 83% sensitivity and 90% specificity while the molecular conformation revealed up to 100% sensitivity and specificity.
The recorded high resistance of the isolated Salmonella reflects a serious problem attributed to the extensive use of the antibiotic. Strategies should be enforced for applying strict hygienic and biosecurity measures in poultry farms and reducing the usage of chemical antibiotics in the poultry farms, as well the use of alternatives like Nigella sativa Linn, clay, probiotics, synbiotics, phytobiotics, magnetic water, Tilapia bone, modified egg-shell, activated wheat/rice straw, and Eichhornia Crassipes that produce strong antimicrobial actions and enhance the immunity levels in poultry.
ACKNOWLEDGMENTS
Sincere thanks should be provided to the community services sector of the faculty of veterinary medicine for the help they provided. Thanks to the broiler farms owner and slaughterhouses for their understanding during sample collection in the study period.
Novelty Statement
The study resides the prevalence rates and antimicrobial resistance of Salmonella isolated from chicken farms and slaughterhouses. As well, the study reported a judgment on the hygienic and biosecurity measures installed in the farms and slaughterhouses understudy based on the detected prevalence of Salmonella.
AUTHOR’S CONTRIBUTION
ESS designed the experimental design, participated in the execution of the bacteriological analysis experiment, and took part in the writing of the manuscript. MHG collected the chicken samples, participated in the execution of the bacteriological analysis experiment, and took a part in the writing of the manuscript. HME supervised the laboratory study and took a part in the writing of the manuscript.
Conflict of interest
The authors have declared no conflict of interest.
REFERENCES
Abd El-Aziz DM (2013). Detection of Salmonella typhimurium in retail chicken meat and chicken giblets. Asian Pac. J. Trop. Biomed., 3(9): 678-681. https://doi.org/10.1016/S2221-1691(13)60138-0
Al-Ansaria MM, Aljubalia MM, Somily AM, Albarrag AM, Masood A (2021). Isolation and molecular characterization of multidrug-resistant Salmonella enterica serovars. J. Infect. Publ. Health, Available online 14 October 2021. https://doi.org/10.1016/j.jiph.2021.10.011
Alsultan MA, Alhammadi MA, Hemida MG (2019). Infectious bronchitis virus from chickens in Al-Hasa, Saudi Arabia 2015-2016. Vet. World, 12: 424–433. https://doi.org/10.14202/vetworld.2019.424-433
Althaus D, Zweifel C, Stephan R (2017). Analysis of a poultry slaughter process: Influence of process stages on the microbiological contamination of broiler carcasses. Ital. J. Food Saf., 6(4): 7097. https://doi.org/10.4081/ijfs.2017.7097
American Public Health Association, American Water Works Association, Water Environment Federation (2017). Standard methods for the examination of water and wastewater. By E.W. Rice, R.B. Baird, A.D. Eaton, American Water Work Association Publications 23rd Ed, Washington D.C.
Antunes P, Mourão J, Campos J, Peixe L (2016). Salmonellosis: the role of poultry meat. Clin. Microbiol. Infect., 22(2): 110-121. https://doi.org/10.1016/j.cmi.2015.12.004
Armwood BT, Rieth A, Baldwin L, Roney CS, Barbieri NL, Logue CM (2019). Assessing the Ability of maternal antibodies to protect broiler chicks against colonization by Salmonella heidelberg. Avian Dis., 63: 289–293. https://doi.org/10.1637/11970-091218-Reg.1
Attia YA, Al-Harthi MA, El-Shafey AS, Rehab YA, Kim WK (2017). Enhancing tolerance of broiler chickens to heat stress by supplementation with vitamin E, Vitamin C and/or Probiotics. Annal. Anim. Sci., 17: 1–15. https://doi.org/10.1515/aoas-2017-0012
Bénard F, Barkun AN, Martel M, Renteln D von (2018). Systematic review of colorectal cancer screening guidelines for average-risk adults: Summarizing the current global recommendations. World J. Gastroenterol., 24(1): 124–138. https://doi.org/10.3748/wjg.v24.i1.124
Braden CR (2006). Salmonella enterica serotype enteritidis and eggs: A national epidemic in the United States. Clin. Infect. Dis., 43(4): 512-517. https://doi.org/10.1086/505973
Cavani C, Petracci M, Trocino A, Xiccato G (2009). Advances in research on poultry and rabbit meat quality. Ital. J. Anim. Sci., 8: 741-750. https://doi.org/10.4081/ijas.2009.s2.741
Centers for Disease Control and Prevention. (2020). Salmonella Page. [(Accessed on 1 December 2020)]; Available online: https://www.cdc.gov/salmonella/index.html#:~:text=CDCestimatesSalmonellabacteriacause,%2Cfever%2Candstomachcramps.
Clinical and Laboratory Standards Institute (2002). Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals. approved standard, 2nd ed. NCCLS Document M31-A2. Clinical and Laboratory Standards Institute, Wayne, PA.
Collins DL, Holmes CJ, Peters TM, Evans AC (1995). Automatic 3-D model-based neuroanatomical segmentation. Hum. Brain Mapp., 3(3): 190-208. https://doi.org/10.1002/hbm.460030304
Cota JB, Silva VFD, Chambel L, Veloso MG, Vieira-Pinto M, Oliveira M (2019). Pheno and genotyping of Salmonella from slaughtered pigs in a Portuguese abattoir reveal differential persistence ability. Vet Microbiol., 239:108457. https://doi.org/10.1016/j.vetmic.2019.108457
Davanzo EFA, dos-Santos RL, Castro VHL, Palma JM, Pribul BR, Dallago BSL, Fuga B, Medeiros M, Almeida SST, de-Costa HMB, Rodrigues DDP, Lincopan N, Perecmanis S, Patricia A (2021). Molecular characterization of Salmonella spp. and Listeria monocytogenes strains from biofilms in cattle and poultry slaughterhouses located in the federal District and State of Goiás, Brazil. PLoS One, 16(11): e0259687. https://doi.org/10.1371/journal.pone.0259687
Delpont M, Guinat C, Guérin JL, Le Leu E, Vaillancourt JP, Paul MC (2021). Biosecurity measures in French poultry farms are associated with farm type and location. Prev. Vet. Med., 195: 105466. https://doi.org/10.1016/j.prevetmed.2021.105466
Delpont M, Racicot M, Durivage A, Fornili L, Guérin JL, Vaillancourt JP, Paul MC (2021). Determinants of biosecurity practices in French duck farms after a H5N8 Highly Pathogenic Avian Influenza epidemic: The effect of farmer knowledge, attitudes and personality traits. Transbound. Emerg. Dis., 68(1): 51-61. https://doi.org/10.1111/tbed.13462
Donado-Godoy P, Clavij V, Leo M, Tafur MA, Gonzales S, Hume M, Alali W, Walls I, Wong DMA, Doyle MP (2012). Prevalence of Salmonella on retail broiler chicken meat carcasses in Colombia. J. Food Prot., 75(6): 1134-1138. https://doi.org/10.4315/0362-028X.JFP-11-513
Elshebrawy HA, Mahros MA, Abd-Elghany SM, Elgazzar MM, Hayashidani H, Sallam KI (2021). Prevalence and molecular characterization of multidrug-resistant and β-lactamase producing Salmonella enterica serovars isolated from duck, pigeon, and quail carcasses in Mansoura, Egypt. LWT, 149 (September 2021): 111834. https://doi.org/10.1016/j.lwt.2021.111834
Fàbrega A, Vila J (2013). Salmonella enterica serovar typhimurium skills to succeed in the host: Virulence and regulation. Clin. Microbiol. Rev., 26(2): 308–341. https://doi.org/10.1128/CMR.00066-12
FAO Statistics (2020). Available online at: http://fenix.fao.org/faostat/internal/en/#home, (accessed May 28, 2020).
Gand M, Mattheus W, Saltykova A, Roosens N, Dierick K, Marchal K, De Keersmaecker SCJ, Bertrand S (2019). Development of a real-time PCR method for the genoserotyping of Salmonella paratyphi B variant Java. Appl. Microbiol. Biotechnol., 103(12): 4987-4996. https://doi.org/10.1007/s00253-019-09854-4
Hafez HM (2005). Governmental regulations and concepts behind eradication and control of some important poultry diseases. World Poult. Sci. J., 61: 569–582. https://doi.org/10.1079/WPS200571
Herigstad B, Hamilton M, Heersink J (2001). How to optimize the drop plate method for enumerating bacteria. J. Microbiol. Meth., 44(2): 121-129. https://doi.org/10.1016/S0167-7012(00)00241-4
Hyeon JY, Chon JW, Hwang IG, Kwak HS, Kim MS, Kim SK, Choi IS, Song CS, Park C, Seo KH (2011). Prevalence, antibiotic resistance, and molecular characterization of Salmonella serovars in retail meat products. J. Food Prot., 74(1): 161-166. https://doi.org/10.4315/0362-028X.JFP-10-327
Johansson MHK, Bortolaia V, Tansirichaiya S, Aarestrup FM, Roberts AP, Petersen TN (2021). Detection of mobile genetic elements associated with antibiotic resistance in Salmonella enterica using a newly developed web tool: Mobile element finder. J. Antimicrob. Chemother., 76(1): 101–109. https://doi.org/10.1093/jac/dkaa390
Kagambega A, Hiott LM, Boyle DS, McMillan EA, Sharma P, Gupta SK, Ramadan H, Cho Sohyun, Humayoun SB, Woodley TA, Barro N, Jackson CR, Frye JG (2021). Serotyping of sub-Saharan Africa Salmonella strains isolated from poultry feces using multiplex PCR and whole genome sequencing. Microbiology, 21: 29. https://doi.org/10.1186/s12866-021-02085-6
Kim SK, Lee JH (2016). Biofilm modeling systems. Korean J. Microbiol., 52(2): 125-139. https://doi.org/10.7845/kjm.2016.6027
Kloska F, Beyerbach M, Klein G (2017). Infection dynamics and antimicrobial resistance profile of Salmonella paratyphi B d-tartrate positive (Java) in a persistently infected broiler barn. Int. J. Environ. Res. Publ. Health, 14(1): 101. https://doi.org/10.3390/ijerph14010101
Le Bouquin S, Allain V, Rouxel S, Petetin I, Picherot M, Michel V, Chemaly M (2010). Prevalence and risk factors for Salmonella spp. contamination in French broiler-chicken flocks at the end of the rearing period. Prev. Vet. Med., 97(3-4): 245-251. https://doi.org/10.1016/j.prevetmed.2010.09.014
Mandal RK, Kwon YM (2017). Global screening of Salmonella enterica serovar typhimurium genes for desiccation survival. Front. Microbiol., 8: 1723. https://doi.org/10.3389/fmicb.2017.01723
Matsui K, Nakazawa C, Khin TMM, Iwabuchi E, Asai T, Ishihara K (2021). Molecular characteristics and antimicrobial resistance of Salmonella enterica Serovar Schwarzengrund from chicken meat in Japan. Antibiotics, 10: 1336. https://doi.org/10.3390/antibiotics10111336
McWhorter AR, Chousalkar KK (2019). From hatch to egg grading: Monitoring of Salmonella shedding in free-range egg production systems. Vet. Res., 2019: 50-58. https://doi.org/10.1186/s13567-019-0677-4
Mir IA, Wani SA, Hussain I, Qureshi SD, Bhat MA, Nishikawa Y (2010). Molecular epidemiology and in vitro antimicrobial susceptibility of Salmonella isolated from poultry in Kashmir. Rev. Sci. Tech. (Int. Off. Epizoot.), 29(3): 677-686. https://doi.org/10.20506/rst.29.3.2011
Morishita TY, Derksen T (2021). Biosecurity chapter 5. In backyard poultry medicine and surgery: A guide for veterinary practitioners, 2nd ed. Wiley online Library. https://doi.org/10.1002/9781119511816.ch5
Murray PR, Rosenthal KS, Pfaller MA (2015). Medical microbiology, 8th Edition, Elsevier health sciences, Philadelphia, PA, USA.
National Committee for Clinical Laboratory Standards (1990). Performance Standards for Antimicrobial Disk Susceptibility Tests. 4th Ed, NCCLS Document MZ-A4. Villanova, PA 19085, U.S.A. file:///C:/Users/Al%20Badr/Downloads/01-CLSI-M02-A11-2012.pdf
Obe T, Nannapanen R, Schilling W, Kiess A (2021). Antimicrobial tolerance, biofilm formation, and molecular characterization of Salmonella isolates from poultry processing equipment. J. Appl. Poult. Res., 30(4): 100195. https://doi.org/10.1016/j.japr.2021.100195
Olivera SD, Rodenbusch CR, Ce MC, Rocha SLS, Canal CW (2003). Evaluation of selective and non-selective enrichment PCR procedures for Salmonella detection. Lett. Appl. Microbiol., 36: 217-221. https://doi.org/10.1046/j.1472-765X.2003.01294.x
Otte J, Rushton J, Rukambile E, Alders RG (2021). Biosecurity in village and other free-range poultry trying to square the circle? Front. Vet. Sci., 8: 678419. https://doi.org/10.3389/fvets.2021.678419
Powers DMW (2011). Evaluation: From precision, recall and F-measure to ROC, informedness, markedness and correlation. J. Mach. Learn. Technol., 2(1): 37–63.
Pulford CV, Perez-Sepulveda BM, Canals R, Bevington JA, Bengtsson RJ, Wenner N, Rodwell EV, Kumwenda B, Zhu X, Bennett RJ, Stenhouse GE, De Silva PM, Webster HJ, Bengoechea JA, Dumigan A, Tran-Dien A, Prakash R, Banda HC, Alufandika L, Mautanga MP, Bowers-Barnard A, Beliavskaia AY, Predeus AV, Rowe WPM, Darby AC, Hall N, Weill FX, Gordon MA, Feasey NA, Baker KS, Hinton JCD (2021). Stepwise evolution of Salmonella Typhimurium ST313 causing bloodstream infection in Africa. Nat. Microbiol., 6: 327–338. https://doi.org/10.1038/s41564-020-00836-1
Rodriguez J, Rondón I, Verjan N (2015). Serotypes of Salmonella in broiler carcasses marketed at Ibague, Colombia. Rev. Bras. Ciência Avícola., 17: 545–552. https://doi.org/10.1590/1516-635X1704545-552
Rodríguez-Hernández R, Bernal JF, Cifuentes JF, Fandiño LC, Herrera-Sánchez MP, Rondón-Barragán L, Garcia NV (2021). Prevalence and molecular characterization of Salmonella isolated from broiler farms at the Tolima Region-Colombia. Animals, 11(4): 970. https://doi.org/10.3390/ani11040970
Rogers AWL, Tsolis RM, Bäumler AJ (2021). Salmonella versus the microbiome. https://doi.org/10.1128/MMBR.00027-19
RosenbergYehezkelHoffmanMattioliFremderBen-AroshVainmanNissaniHen-AviviBrennerItkinOhanaBen-MosheAvrahamHost succinate is an activation signal for Salmonella virulence during intracellular infection. Science, 371(6527): 400-405. https://doi.org/10.1126/science.aba8026
Ryan MP, O’Dwyer J, Adley C (2017). Evaluation of the Complex Nomenclature of the Clinically and Veterinary Significant Pathogen Salmonella. BioMed. Res. Int., 2017: 1–6. https://doi.org/10.1155/2017/3782182
Sambrook J, Fritscgh EF, Mentiates (1989). Molecular coloning. A laboratory manual. Vol 1., Cold spring Harbor Laboratotry press, New York. https://www.cshlpress.com/pdf/sample/2013/MC4/MC4FM.pdf
Schneid ADS, Rodrigues KL, Chemello D, Tondo EC, Ayub MAZ, Aleixo JAG (2006). Evaluation of an indirect ELISA for the d of Salmonella in chicken meat. Braz. J. Microbiol., 37: 350-355. https://doi.org/10.1590/S1517-83822006000300027
Schweitzer PM, Susta L, Varga C, Brash M, Guerin MT (2021). Demographic, husbandry, and biosecurity factors associated with the presence of campylobacter spp. in small poultry flocks in Ontario, Canada. Pathogens, 10(11): 1471. https://doi.org/10.3390/pathogens10111471
Sheela R, Babu U, Mu J, Elankumaran S, Bautista D, Raybourne R, Heckert RA, Song W (2003). Immune responses against Salmonella enterica serovar enteritidis infection in virally immunosuppressed chickens. Clin. Vaccine Immunol., 10: 670-679. https://doi.org/10.1128/CDLI.10.4.670-679.2003
Soliman ES, Moawed SA, Ziaan AMG (2016). Assessing cleaning and disinfection regime in a slaughterhouse against carcasses contamination. Adv. Anim. Vet. Sci., 4(9): 449-457. https://doi.org/10.14737/journal.aavs/2016/4.9.449.457
SPSS (2016). Statistical packages of social sciences. Version 21 for windows. SPSS. Inc. USA. https://www.ibm.com/support/pages/spss-statistics-210-available-download
Su LH, Chiu CH (2007). Salmonella: Clinical importance and evolution of nomenclature. Chang Gung Med. J., 30(3): 210-219.
Thai TH, Yamaguchi R (2012). Molecular characterization of antibiotic-resistant salmonella isolates from retail meat from markets in northern Vietnam. J. Food Prot., 75(9): 1709-1714. https://doi.org/10.4315/0362-028X.12-101
Uddin MB, Hossain SMB, Hasan M, Alam MN, Debnath M, Begum R, Roy S, Ahmed Harun-Al-Rashid A, Chowdhury MSR, Rahman MM, Hossain MM, Elahi F, Chowdhury MYE, Järhult JD, El Zowalaty ME, Ahmed SSU (2021). Multidrug antimicrobial resistance and molecular detection of mcr-1 gene in Salmonella species isolated from chicken. Animals, 11(1): 206. https://doi.org/10.3390/ani11010206
Voss-Rech D, Kramer B, Silva VS, Rebelatto R, Abreu PG, Coldebella A, Vaz CSL (2019). Longitudinal study reveals persistent environmental Salmonella heidelberg in Brazilian broiler farms. Vet. Microbiol., 233: 118–123. https://doi.org/10.1016/j.vetmic.2019.04.004
Yildirim Y, Gonulalan Z, Pamuk S, Ertas N (2011). Incidence and antibiotic resistance of Salmonella spp. on raw chicken carcasses. Food Res. Int., 44(3): 725-728. https://doi.org/10.1016/j.foodres.2010.12.040
Yu L, Yang X, Zhang J, Yang S, Zhang S, Chen M, Xue L, Ding Y, Zeng H, Gu Q, Zhang Y, Wei X, Wang J, Wu Q (2021). Molecular characterisation of antimicrobial resistance determinants and class 1 integrons of Salmonella enterica subsp. Enterica serotype Enteritidis strains from retail food in China. Food Contr., 128: 108191. https://doi.org/10.1016/j.foodcont.2021.108191
To share on other social networks, click on any share button. What are these?





