Preliminary Behavioral, Pathological and Transcriptome Studies of Tree Shrew: Evidence for a Complementary Small-Animal Alzheimer's Disease Model
Preliminary Behavioral, Pathological and Transcriptome Studies of Tree Shrew: Evidence for a Complementary Small-Animal Alzheimer’s Disease Model
Yi-qiang Ouyang1, Zhen-xin Liang1, Shi-wen Huang2, Ying Zhang3, Shao-shi Luo1, Jin-ning Liang1, Jia-fu Li1, Yu He4 and Song-chao Guo1,*
1Laboratory Animal Center, Guangxi Medical University, Nanning 530021, Guangxi, China
2Guangxi Institute for the Prevention and Treatment of Occupational Diseases, Nanning 530021, Guangxi, China
3Biological Targeting Therapy & Diagnosis Center of Guangxi Medical University, Nanning 530021, Guangxi, China
4The First Affiliated Hospital of Guangxi Medical University, Nanning 530021, Guangxi, China
ABSTRACT
Most small-animal studies of Alzheimer’s disease (AD) involve rats and mice, yet the tree shrew (Tupaia belangeri) is in many respects more similar to primates than rodents are. To demonstrate the feasibility and usefulness of using the tree shrew as an AD model, we examined how well the model recapitulates features of the disease. Injecting animals intraperitoneally with d-galactose to induce acute senility, and then injecting them with β amyloid fragment 1-42 and ibotenic acid into the bilateral hippocampus led to significant learning and memory deficits in the Morris water maze test. Pathology analysis of treated animals showed obvious gliosis and neurofibrillary tangles in the cerebral hippocampal area. These results suggest that the tree shrew can recapitulate the major features of AD pathogenesis. To provide the basis for further studies with this new animal model, we used high-throughput sequencing to analyze changes in the hippocampal transcriptome induced by injection of the three AD agents. These results open the door to new small-animal studies of AD that complement and extend studies in rodents and primates.
Article Information
Received 5 October 2016
Revised 12 December 2016
Accepted 20 January 2017
Available online 14 June 2017
Authors’ Contribution
ScG conceived and designed the study. YqO and ZxL performed the experimental work and recorded observation. SsL, JnL and JfL fed the tree shrews. YqO, SwH and YH analyzed the data. YqO and YZ wrote the article.
Key words
Tree shrew, Alzheimer’s disease, Animal model, High-throughput sequencing.
DOI: http://dx.doi.org/10.17582/journal.pjz/2017.49.4.1231.1241
* Corresponding author: 2433164518@qq.com
0030-9923/2017/0004-1231 $ 9.00/0
Copyright 2017 Zoological Society of Pakistan
INTRODUCTION
Alzheimer’s Disease (AD) is a devastating neurodegenerative disease that poses an increasing health burden worldwide, and a cure remains out of sight (Ikram and DeCarli, 2012). Since the disease currently can be diagnosed definitively only after death, studies in animal models of AD provide the only way to obtain in vivo information about disease onset and progression. Primates such as macaque and rhesus monkeys provide good model systems (Darusman et al., 2014; Kimura et al., 2003; Okabayashi et al., 2015; Pahl et al., 2013; Park et al., 2015; Vardigan et al., 2015), but such studies require significant time, expense and facilities. In addition, ethical concerns restrict the size of such studies to relatively few animals. To avoid these disadvantages, many researchers rely on rat and mouse models (Bader Lange et al., 2010; Cummings et al., 2015; Li et al., 2015; Martino Adami et al., 2015; Zahedi et al., 2015), but neocortical structures differ between rodents and humans. This may help explain why most drugs developed in rodent studies fail to show satisfactory efficacy in humans. While a few non-primate animal models of AD have been developed, including cats, dogs, pigs and chickens (Neus Bosch et al., 2015; Vite and Head, 2014; Zhang et al., 2015; Sondergaard et al., 2012; Yan et al., 2015), none of these models perfectly recapitulates all pathological changes in AD. Thus new animal models are needed, particularly small animals that may be more similar to humans than rodents are.
The tree shrew, Tupaia belangeri, which belongs to the Tupaiidae of Scandentia and is found primarily in China, Vietnam and Burma, attracted the attention of biomedical researchers when it was initially thought to be the only non-primate that could be infected by hepatitis virus. Genetic studies indicate that the tree shrew is more closely related to primates than rodents are (Fan et al., 2013; Song et al., 2012; Lind-Blod-Toh et al., 2011; Hallström and Janke, 2008), with a larger brain and more advanced nervous system than those of rats. Much is already known about tree shrew biology, and the animal has gradually attracted attention for its potential as a model of human nervous system diseases. It has not yet been analyzed in detail for its usefulness as a model of AD.
If the tree shrew can be demonstrated to be a good AD model, it would provide numerous advantages over primate and rodent models. Its smallness and high fertility make it a much more efficient and cost-effective experimental system than primates. Since tree shrews and rats are similar in overall size and head shape, the same stereotaxic apparatus for rats can be used for tree shrews. To probe the feasibility and usefulness of the tree shrew as an AD model, we examined the behavior and pathology of animals treated with the combination of d-galactose, β amyloid fragment 1-42 and ibotenic acid. We assessed the behavioral and pathology properties of the treated animals in the same way that rat models of AD are assessed. We also screened the hippocampal transcriptome for changes in gene expression resulting from experimental AD induction. The results of this study should help lay the foundations for using the tree shrew as a small-animal AD model to complement studies in rodents and primates.
MATERIALS AND METHODS
Animals
Male tree shrews (n = 30) aged three months and weighing 120±10 g were obtained from the Kunming Medical University Laboratory Animal Center (animal license No. SCXK Yunnan 2013-0002). All animal manipulations and experiments were carried out at the Laboratory Animal Center of Guangxi Medical University (animal license no. SYXK Guangxi 2014-0002).
Reagents and instruments
β amyloid 1-42 peptide (Aβ1-42), ibotenic acid (Ibo), and d-galactose (d-gal) were purchased from Sigma (St.Louis, USA). Aβ1-42 was dissolved to a concentration of 2 g/L in sterilized saline and incubated for 72 h at 37 °C to form condensed state. The following major instruments were used: stereotactic frame (RWD68002, Shenzhen RWD Life Science, Shenzhen, China), Morris water maze (built in-house by Guangxi Medical University), microsyringe (Shanghai Anting Microsyringe Factory, Shanghai, China); and electronic balance (ML204, Mettler-Toledo, Shanghai, China).
Animal group allocation
The 30 tree shrews were divided randomly into saline, blank and treatment groups (n = 10 in each). Blank animals did not undergo any surgical procedures, while the treatment group was injected with three AD-inducing agents as described for rats (Jia et al., 2015). Saline animals underwent the same procedures as the treatment group, except that saline was used instead of AD-inducing agents.
Establishment of AD model
Animals in the treatment group received daily intraperitoneal injections of d-Gal (50 mg/kg−1) for 30 days in order to induce senility. On day 31, animals were anesthetized by intraperitoneal injection of 1% sodium pentobarbital (2 mL/kg-1) and immobilized in a rat stereotactic frame. The skin was cut along the central parietal scalp to expose the sagittal suture. Beginning from the intersection of the sagittal suture and biauricular line, the skull was drilled 3.3 mm forward at a distance of 5 mm on both sides. Then the stereotactic instrument was used to stab the microsyringe needle vertically through the opening to a depth of 10 mm into the brain. The needle was then inserted slowly at a constant speed for 5 min until the needle tip arrived at the hippocampus. A mixture of 10 μg Aβ1-42 and 10 μg Ibo dissolved in 5 μL of saline was injected slowly over 15 min. The needle was left in place for 10 min to prevent fluid pull-back, then it was pulled out slowly; the incision was sutured and sterilized with 75% ethyl alcohol. Animals in the saline group were treated in the same way, except that saline was used instead of d-Gal, Aβ1-42 or Ibo.
The appearance, eating and drinking patterns, and locomotor activity of animals was carefully monitored after intracerebral injection. At 4 weeks after injection, behavioral tests were performed. Finally, animals in the treatment and saline groups were anesthetized by intramuscular injection of ketamine (100 mg/kg-1) at 25°C in the laboratory. Brain tissue was immediately removed and immersed in liquid nitrogen, then stored at
-80°C.
Morris water maze
The learning and memory abilities of treated animals were compared with those in the saline and blank groups using the classical Morris water maze. The Morris water maze system comprised a circular basin maze filled with water (diameter, 150 cm; depth, 60 cm), a platform under the water (height, 30 cm; radius, 5 cm) and a video analysis system for on-line digital image acquisition and off-line image analysis. Images were collected using a camera positioned 3 m above the center of the pool. The pool was evenly divided into four quadrants (NE, SE, SW and NW), which were conspicuously labeled in the middle of the wall within each quadrant. The platform was made of pure black plastic and placed in the middle of the desired quadrant. The water level in the pool was 0.5 cm above the platform. The water in the basin was colored white using milk powder to render the platform invisible. The water was maintained at a temperature of 21±0.5°C. The lighting in the room was maintained constant, with no direct illumination of the pool, and noise in the room was kept to a minimum.
During initial adaptation, tree shrews were put in the water with their head facing the pool wall, and they were guided to the underwater platform. All animals were placed randomly at starting points positioned at the north, south, east and west ends of the pool. The time required for the animal to find the platform was recorded; if the time exceeded 75 sec, the animals were guided to the platform and forced to stay there for 10 sec. Animals were trained in this way twice daily for seven consecutive days. Only those who swam well and found the platform easily by themselves were used for behavioral tests (see next section). After each training run, animals were immediately dried with towels and an electric hair dryer to prevent hypothermia.
Navigation tests, each lasting 75 sec, were carried out between 8:30 and 10:00 a.m. on four consecutive days. The animal was gently placed in the pool with its head facing the pool wall randomly at one of four positions (north, south, east and west). The time needed to find and climb onto the platform was recorded, allowing calculation of escape latency and swimming speed. Animals that found the platform were forced to stay there for 10 sec in order to adapt to the platform and the surroundings. If animals did not find the platform within 75 sec, the computer stopped tracking and recorded the escape latency as 75 sec, and the experimenter guided the animals to the platform, where they were forced to stay for 15 sec. After each test, animals were immediately dried with towels and an electric hair dryer to prevent hypothermia.
On the second day after navigation tests, a so-called “probe test” was conducted in which the platform was removed and animals were ramdonly placed at the four positions in the pool (north, south, east and west) facing the pool wall. The animals were allowed to swim for 90 sec, and the frequency with which they crossed over the area where the platform had previously been located was recorded. This frequency provided an index of spatial memory.
Pathology analysis
After behavioral testing, animals were anesthetized by intraperitoneal injection of 1% sodium pentobarbital (0.15 mL/100 g), and then fixed as follows. The chest was opened from the xyphoid, the animal was intubated into the aorta via the left ventricular, and rapidly perfused with 100 mL saline to flush out blood. Then the animal was perfused with 500 mL 4% paraformaldehyde, and brain tissue was removed and fixed immediately in 4% neutral formalin. Tissue was sectioned, embedded in paraffin and stained with hematoxylin-eosin for morphological examination. Sections were impregnated with Bieloschsky silver to detect neurofibrillary tangles.
High-throughput sequencing of the hippocampal transcriptome
After behavioral testing, animals in the treatment and saline groups were anesthetized by intramuscular injection of ketamine (100 mg/kg) at 25 °C in the laboratory. Brain tissue was removed, flash-frozen in liquid nitrogen, and stored at -80 °C. Hippocampal tissue was isolated from frozen brain tissue (see previous section) and total RNA (10 μg) was extracted using the RNeasy Micro kit (Qiagen, cat. No. 74004). This RNA was converted into a cDNA library using the RNeasy Micro kit (Cat#74004, QIAGEN) according to the manufacturer’s instructions.
High-throughput sequencing was conducted using the platform of Shanghai Biotechnology and the HiSeq 2500 sequencer (Illumina). Paired-end software (Illumina) was used to perform 2 x 100-nt multiplexing. Routine methods were used to analyze the RNA-seq transcriptome of tree shrews in the treatment and saline groups, including preprocessing, fragment assembly, database annotation, differential expression analysis, gene ontology (GO) function classification, Cluster of Ortholog Genes (COG) classification, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway mapping, and analysis of GO functional classes and KEGG pathways enriched for differentially expressed genes. Unqualified reads containing sequencing primers were removed, and fragments were spliced de novo using the scaffolding contig algorithm of CLC Genomics Workbench 6.0.4 (Brautigam et al., 2011; Garg et al., 2011; Jia et al., 2015) and the parameters Word-size=45 and Minimum contig length>=300. The spliced sequence at this stage, termed the primary UniGene sequence, was spliced again using CAP3 EST software, generating a final UniGene sequence set. The first splicing was performed with loose splicing parameters; the secondary splicing, with strict parameters.
The final UniGene spliced sequence set was translated into protein using trEMBL+swissprot and compared with the UniProt library (release 2013.04) using BLASTX. Sequence annotation and comparison were performed using a significance threshold of E <10-5.
With this final UniGene spliced sequence set serving as reference, reads of each sample were mapped to measure UniGene coverage of each sample. The expression level of reads was quantitated using the following formula for reads per kb per million (RPKM):
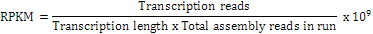
Where, transcription reads is the number of reads covering the entire UniGene; transcription length is the length of the UniGene; total assembly reads in run is the total number of reads involved in splicing the sample.
The RPKM of each sample was determined and compared with the total expression level as an internal standard; the difference was assessed for significance using Fisher’s test. The DEGseq analysis package was used to determine genes differentially expressed between the treatment and saline groups, using the screening criteria of P < 0.05 and fold-change ≥1.5.
Functional classification of differentially expressed hippocampal genes
All differentially expressed hippocampal genes were mapped to GO classification terms, the number of genes in each term was counted, and then the terms enriched in differentially expressed genes compared to the whole genome were identified using a hypergeometric test:
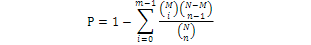
Where, N is the number of genes with GO annotations; n, the number of differentially expressed genes within N; M, the number of all genes annotated as specific GO term; and m, the number of differentially expressed genes annotated as specific GO term. GO terms enriched in differentially expressed genes were those associated with a Bonferroni-corrected P ≤ 0.05. GO functions were determined by applying the blast2GO algorithm to the UniProt annotation results. Sequences were assigned to one of three GO categories: molecular function, cellular components, and biological processes.
COG functions were determined by comparing UniGenes and the CDD library using rpstblastn. This algorithm predicts COG functional classification based on the five best results for a threshold of E < 10-5. KEGG mapping of the final UniGene spliced sequence was performed using the online KEGG KAAS tool (http://www.genome.jp/tools/kaas/).
Statistical analysis
All results were expressed as mean±SD and statistical tests were performed using SPSS 19.0 (IBM, Chicago, IL, USA). Pair-wise comparisons were assessed for significance using the least-squares difference tests if the data showed homogeneous variance; otherwise, the Games-Howell test was used. Comparisons among more than two groups were assessed using one-way analysis of variance (ANOVA). A two-sided P < 0.05 was defined as the threshold of significance.
RESULTS
Escape latency during training in the Morris water maze
During the 7-day training period, the blank and saline groups showed similar training profiles, with escape latency gradually decreasing to similar extents and at similar rates in the two groups (Fig. 1). In contrast, the treatment group showed significantly longer escape latency than the blank group from day 3 onwards, and longer latency than the saline group from day 4 onwards.
Table I.- Performance of tree shrews in the probe test.
|
Group |
Crossings of target quadrant | Swimming speed (cm/s) |
| Treatment |
1.33±1.32ab |
15.21±6.75 |
| Saline | 3.15±1.98 | 14.87±7.56 |
| Blank | 4.11±1.95 |
15.37±6.74 |
Values are mean ± SD for each group of 10 animals.a P < 0.05 vs. the blank group; b P < 0.05 vs. the saline group.
Probe test in the Morris water maze
All three animal groups showed similar swimming speed. Both the blank and saline groups showed a similar tendency to search within the quadrant where the platform had been located earlier. In contrast, the treatment group tended to search at random (Table I).
Pathology analysis in the hippocampus
Hematoxylin-eosin staining of hippocampal sections from tree shrew revealed no apparent histopathology in blank and saline animals, but gliosis in treated animals (Fig. 2). Similarly, Bieloschsky silver impregnation revealed no clear pathology in the blank and saline groups, but neurofibrillary tangles in the treatment group (Fig. 3).
Differential expression of hippocampus genes after AD induction
A total of 44,430,000 clean reads were obtained and spliced to generate the initial UniGene sequence set. ESTs were switched together to generate the final UniGene sequence set, comprising 104,504,006 bp and 99,463 counts. The average sequence length was 1,050 bp, and N50 was 1,695 bp (Table II). Analysis of the length distribution in the final UniGene sequence set (Fig. 4) showed that 33,576 sequences were 400-600 bp long, accounting for 33.8% of all UniGenes; 21,353 were 200-400 bp, accounting for 21.5%; and 12,862 were 400-600 bp long, accounting for 12.9%.
Table II.- Statistics of UniGene sequence assembly of the tree shrew hippocampal transcriptome.
| Parameters | Contigs |
Initial UniGene |
Final UniGene |
| Counts (n) | 116,654 | 100,872 | 99,463 |
| Total length (bp) | 104,313,695 | 104,610,822 | 104,504,006 |
| N25 (bp) | 3,229 | 3,945 | 4,001 |
| N50 (bp) | 1,384 | 1,650 | 1,695 |
| N75 (bp) | 567 | 633 | 645 |
| Average length (bp) | 894 | 1,037 | 1,050 |
| Max length (bp) | 21,469 | 23,750 |
23,750 |
Functional classification of differentially expressed hippocampal genes
After translating the UniGene nucleotide sequences into protein sequences and comparing them with the UniProt library, a total of 63,708 UniGene sequences were annotated using the GO function (Fig. 7).
To further uncover the range of hippocampal genes expressed in tree shrew, we classified the final UniGene sequence set in terms of COG functional classification. In the end, we were able to assign 19,305 sequences to a total of 25 COG functional classes (Fig. 5).
After functionally classifying the genes identified from the tree shrew hippocampal transcriptome, we analyzed which gene functions were under- and overexpressed following experimental induction of AD using d-Gal, Aβ1-42 and Ibo. We plotted relative transcript levels in the saline group against levels in the treatment group (Fig. 6). This analysis identified a total of 125 genes differentially expressed between the saline and treatment groups, of which 74 were up-regulated in the treatment group and 51 were down-regulated.
Next we clustered the differentially expressed genes according to their GO functional class (Fig. 8). These results suggest that AD in tree shrews is associated with up-regulation of numerous cellular processes in the hippocampus, including glycolysis, toxin response, carbohydrate processing, cellular projections, as well as with activity of 6-phosphofructokinase and M-band.
To complement the GO and COG functional classification of differentially expressed hippocampal genes, we also analyzed up- and down-regulated genes based on KEGG pathways (Fig. 9).
The highest proportion of differentially expressed genes was involved in retinol metabolism, followed by fructose, mannose and galactose metabolism. These results suggest that glucose and retinol metabolic pathways are dysregulated in the tree shrew model of AD.
The parallel analysis of how differentially expressed hippocampal genes distributed across GO and KEGG categories led us to identify several UniGenes up- or down-regulated following AD induction that are likely to be associated with multiple metabolic pathways. These genes included hexokinase 1(HK1), major histocompatibility complex, class I, B(HLA-B), heat shock protein family A (Hsp70) member 1A(HSPA1A), phosphofructokinase, platelet(PFKP), gamma-aminobutyric acid (GABA) A receptor(Gabrd), period circadian clock 3 (PER3), adaptor-related protein complex 2, beta 1 subunit(AP2B1), heat shock protein family A (Hsp70) member 5(HSPA5), prodynorphin(PDYN). These genes may therefore be candidate genes that drive the onset and/or progression of AD in tree shrew.
DISCUSSION
The present study provides pathology and behavioral evidence that tree shrews injected with d-gal, Aβ1-42, and Ibo can recapitulate several features of AD and may therefore make a good small-animal model for studying onset and progression of the disease. This triple combination of AD-inducing agents has been used in rat models of AD and macaque models (Darusman e al., 2014; Jia et al., 2015); this combination was chosen instead of a single inducing factor in an effort to trigger more of the complex pathology of AD. This tree shrew model, like other damage-based animal models of AD (Schliebs and Arendt, 2011), may provide information complementary to that obtained using aged animal models and transgenic animal models.
Tree shrews in our treatment group showed a significant deficit of learning and memory in Morris water maze tests. Such deficits can occur as a result of brain trauma during the microinjection procedure, but we can exclude this possibility because a group microinjected with saline showed similar performance as a blank group that was not microinjected. Furthermore, AD induction in tree shrew did not affect its swimming speed in water, further arguing against generic brain trauma. Our results suggest that tree shews can show AD-like behavioral symptoms, justifying further work in this area. In addition, blank and saline tree shrews in our probe test behaved similarly to rats (Zahedi et al., 2015), despite the fact that tree shrews have been domesticated for just over 30 years. This highlights the feasibility of using the tree shrew as a complement to rat models of AD.
Pathology examination of tree shrews in our treatment group revealed gliosis and neurofibrillary tangles that were absent from the blank and saline groups. This pathology is consistent with that reported for rat and macaque models (Darusman e al., 2014; Jia et al., 2015) of AD. These features overlap substantially with those of human AD, which include selective deletion of cholinergic neurons and synapses in the frontotemporal lobe and hippocampus, senile plaques, and neurofibrillary tangles (St. George-Hyslop and Petit, 2005). In this way, our pathology studies support the behavioral experiments suggesting that tree
shrew can recapitulate many of the features of human AD, potentially making it a good disease model.
As a first step toward demonstrating the feasibility of identifying genes and proteins involved in AD onset and pathogenesis, we performed high-throughput sequencing of the hippocampal transcriptome and identified numerous genes up- or down-regulated following combination treatment with d-gal, Aβ, and Ibo. These genes participate in a broad array of functions, based on GO and COG classification analyses, suggesting that AD involves the dysregulation of numerous cellular pathways. One intriguing finding is abnormal glucose and retinol metabolism in the treatment group, raising the possibility that this dysregulation causes an inflammatory response in the hippocampus, which may be associated with AD.
CONCLUSION
The intraperitoneal injection of D-Gal results in acute aging. One-time low injection of A β1-42 and Ibo mixture in bilateral hippocampus can result in the formation of SP and NFT in nerve tissues and the declines of tree shrew in learning ability and memory. Tree shrew is therefore an ideal replication AD model. Our findings present several testable hypotheses for future studies examining the potential role of specific genes and metabolic pathways in AD. These results open the door to new small-animal studies of AD that complement and extend studies in rodents and primates.
ACKNOWLEDGEMENTS
National Natural Science Foundation of China (31560612); Guangxi Science and Technology Infrastructure Construction (12-97-23).
Statement of conflict of interest
Authors have declared no conflict of interest.
REFERENCES
Bader Lange, M.L., St Clair, D., Markesbery, W.R., Studzinski, C.M., Murphy, M.P. and Butterfield, D.A., 2010. Age-related loss of phospholipid asymmetry in APP(NLh)/APP(NLh) x PS-1(P264L)/PS-1(P264L) human double mutant knock-in mice: relevance to Alzheimer disease. Neurobiol. Dis., 38: 104-115. https://doi.org/10.1016/j.nbd.2010.01.004
Brautigam, A., Mullick, T., Schliesky, S. and Weber, A.P., 2011. Critical assessment of assembly strategies for non-model species mRNA-Seq data and application of next-generation sequencing to the comparison of C(3) and C(4) species. J. exp. Bot., 62: 3093-3102. https://doi.org/10.1093/jxb/err029
Cummings, D.M., Liu, W., Portelius, E., Bayram, S., Yasvoina, M., Ho, S.H., Smits, H.,Ali, S. S.,Steinberg, R.,Pegasiou, C. M.,James, O. T.,Matarin, M.,Richardson, J. C.,Zetterberg, H.,Blennow, K.,Hardy, J. A.,Salih, D. A. and Edwards, F. A., 2015. First effects of rising amyloid-beta in transgenic mouse brain: synaptic transmission and gene expression. Brain, 138: 1992-2004. https://doi.org/10.1093/brain/awv127
Darusman, H.S., Gjedde, A., Sajuthi, D., Schapiro, S.J., Kalliokoski, O., Kristianingrum, Y.P., Handaryani, E. and Hau, J., 2014. Amyloid beta1-42 and the phoshorylated tau threonine 231 in brains of aged cynomolgus monkeys (Macaca fascicularis). Front Aging Neurosci., 6: 313. https://doi.org/10.3389/fnagi.2014.00313
Fan, Y., Huang, Z.Y., Cao, C.C., Chen, C.S., Chen, Y.X., Fan, D.D., He J., Hou H.L., Hu L., Hu X.T., Jiang X.T., Lai R., Lang Y.S., Liang B., Liao S.G., Mu D., Ma Y.Y., Niu Y.Y., Sun X.Q., Xia J.Q., Xiao J., Xiong Z.Q., Xu L., Yang L., Zhang Y., Zhao W., Zhao X.D., Zheng Y.T., Zhou J.M., Zhu Y.B., Zhang G.J., Wang J. and Yao Y.G., 2013. Genome of the Chinese tree shrew. Nat. Commun., 4: 1426. https://doi.org/10.1038/ncomms2416
Garg, R., Patel, R.K., Tyagi, A.K. and Jain, M., 2011. De novo assembly of chickpea transcriptome using short reads for gene discovery and marker identification. DNA Res., 18: 53-63. https://doi.org/10.1093/dnares/dsq028
Hallström, B.M. and Janke, A., 2008. Resolution among major placental mammal interordinal relationships with genome data imply that speciation influenced their earliest radiations. BMC Evolut. Biol., 8: 162. https://doi.org/10.1186/1471-2148-8-162
Ikram, M.A. and DeCarli, C., 2012. Next frontiers in the genetic epidemiology of Alzheimer’s disease. Eur. J. Epidemiol., 27: 831-836. https://doi.org/10.1007/s10654-012-9742-2
Jia, S., Lu, Z., Gao, Z., An, J., Wu, X., Li, X., Dai X., Zheng Q. and Sun Y., 2015. Chitosan oligosaccharides alleviate cognitive deficits in an amyloid-beta-induced rat model of Alzheimer’s disease. Int. J. Biol. Macromol., https://doi.org/10.1016/j.ijbiomac.2015.11.011
Jia, X.L., Wang, G.L., Xiong, F., Yu, X.R., Xu, Z.S., Wang, F. and Xiong, A.S., 2015. De novo assembly, transcriptome characterization, lignin accumulation, and anatomic characteristics: novel insights into lignin biosynthesis during celery leaf development. Sci. Rep., 5: 8259. https://doi.org/10.1038/srep08259
Kimura, N., Tanemura, K., Nakamura, S-I., Takashima, A., Ono, F., Sakakibaram I., Ishii, Y., Kyuwa, S. and Yoshikawa, Y., 2003. Age-related changes of Alzheimer’s disease-associated proteins in cynomolgus monkey brains. Biochem. biophys. Res. Commun., 310: 303-311. https://doi.org/10.1016/j.bbrc.2003.09.012
Li, H., Kang, T., Qi, B., Kong, L., Jiao, Y., Cao, Y., Zhang, J. and Yang, J., 2015. Neuroprotective effects of ginseng protein on PI3K/Akt signaling pathway in the hippocampus of D-galactose/AlCl inducing rats model of Alzheimer’s disease. J Ethnopharmacol., https://doi.org/10.1016/j.jep.2015.12.020
Lindblad-Toh, K., Garber, M., Zuk, O., Lin, M.F., Parker, B.J., Washietl, S., Kheradpour, P., Ernst, J., Jordan, G., Mauceli, E., Ward, L.D., Lowe, C.B., Holloway, A.K., Clamp, M., Gnerre, S., Alfaldi, J., Beal, K., Chang, J., Clawson, H., Cuff, J., Di, Palma F., Fitzgerald, S., Flicek, P., Guttman, M., Hubisz, M.J., Jaffe, D.B., Jungreis, I., Kent, W.J., Kostka, D., Lara, M., Martins, A.L., Massingham, T., Moltke, I., Raney, B.J., Rasmussen, M.D., Robinson, J., Stark, A., Vilella, A.J., Wen, J., Xie, X., Zody, M.C.; Broad Institute Sequencing Platform and Whole Genome Assembly Team, Baldwin, J., Bloom, T., Chin, C.W., Heiman, D., Nicol, R., Nusbaum, C., Young, S., Wilkinson, J., Worley, K.C., Kovar, C.L., Muzny, D.M., Gibbs, R.A.; Baylor College of Medicine Human Genome Sequencing Center Sequencing Team, Cree, A., Dihn, H.H., Fowler, G., Jhangiani, S., Joshi, V., Lee, S., Lewis, L.R., Nazareth, L.V., Okwuonu, G., Santibanez, J., Warren, W.C., Mardis, E.R., Weinstock, G.M., Wilson, R.K.; Genome Institute at Washington University, Delehaunty, K., Dooling, D., Fronik, C., Fulton, L., Fulton, B., Graves, T., Minx, P., Sodergren, E., Birney, E., Margulies, E.H., Herrero, J., Green, E.D., Haussler, D., Siepel, A., Goldman, N., Pollard, K.S., Pedersen, J.S., Lander, E.S. and Kellis, M., 2011. A high-resolution map of human evolutionary constraint using 29 mammals. Nature, 478: 476-482. https://doi.org/10.1038/nature10530
Martino Adami, P.V., Quijano, C., Magnani, N., Galeano, P., Evelson, P., Cassina, A., Do Carmo, S., Leal, M. C., Castano, E. M., Cuello, A. C. and Morelli, L., 2015. Synaptosomal bioenergetic defects are associated with cognitive impairment in a transgenic rat model of early Alzheimer’s disease. J. Cereb. Blood Flow Metab., https://doi.org/10.1177/0271678X15615132
Neus Bosch, M., Pugliese, M., Andrade, C., Gimeno-Bayon, J., Mahy, N. and Rodriguez, M.J., 2015. Amyloid-beta immunotherapy reduces amyloid plaques and astroglial reaction in aged domestic dogs. Neurodegener Dis., 15: 24-37. https://doi.org/10.1159/000368672
Okabayashi, S., Shimozawa, N., Yasutomi, Y., Yanagisawa, K. and Kimura, N., 2015. Diabetes mellitus accelerates Abeta pathology in brain accompanied by enhanced GAbeta generation in nonhuman primates. PLoS One, 10: e0117362. https://doi.org/10.1371/journal.pone.0117362
Pahl, L., Schubert, S., Skawran, B., Sandbothe, M., Schmidtke, J. and Stuhrmann, M., 2013. 1,25-Dihydroxyvitamin D decreases HTRA1 promoter activity in the rhesus monkey--a plausible explanation for the influence of vitamin D on age-related macular degeneration? Exp. Eye Res., 116:234-239. https://doi.org/10.1016/j.exer.2013.09.012
Park, S.J., Kim, Y.H., Nam, G.H., Choe, S.H., Lee, S.R., Kim, S.U., Kim, J. S., Sim, B. W., Song, B. S., Jeong, K. J., Lee, Y., Park, Y. I., Lee, K. M., Huh, J. W. and Chang, K. T., 2015. Quantitative expression analysis of APP pathway and tau phosphorylation-related genes in the ICV STZ-induced non-human primate model of sporadic Alzheimer’s disease. Int. J. mol. Sci., 16: 2386-23402. https://doi.org/10.3390/ijms16022386
Schliebs, R. and Arendt, T., 2011. The cholinergic system in aging and neuronal degeneration. Behav. Brain Res., 221: 555-563. https://doi.org/10.1016/j.bbr.2010.11.058
Sondergaard, L.V., Ladewig, J., Dagnaes-Hansen, F., Herskin, M.S. and Holm, I.E., 2012. Object recognition as a measure of memory in 1-2 years old transgenic minipigs carrying the APPsw mutation for Alzheimer’s disease. Transgen. Res., 21: 1341-1348. https://doi.org/10.1007/s11248-012-9620-4
Song, S., Liu, L., Edwards, S.V. and Wu, S.Y., 2012. Resolving conflict in eutherian mammal phylogeny using phylogenomics and the multispecies coalescent model. Proc. natl. Acad. Sci. USA., 109: 14942-14947. https://doi.org/10.1073/pnas.1211733109
St George-Hyslop, P.H. and Petit, A., 2005. Molecular biology and genetics of Alzheimer’s disease. Compt. Rend. Biol., 328: 119-130. https://doi.org/10.1016/j.crvi.2004.10.013
Vardigan, J.D., Cannon, C.E., Puri, V., Dancho, M., Koser, A., Wittmann, M., Kuduk S.D., Renger J.J. and Uslaner J.M., 2015. Improved cognition without adverse effects: novel M1 muscarinic potentiator compares favorably to donepezil and xanomeline in rhesus monkey. Psychopharmacology (Berl). 232: 1859-1866. https://doi.org/10.1007/s00213-014-3813-x
Vite, C.H. and Head, E., 2014. Aging in the canine and feline brain. Vet. Clin. N. Am. Small Anim. Pract., 44: 1113-1129. https://doi.org/10.1016/j.cvsm.2014.07.008
Yan, Q., Weyn-Vanhentenryck, S.M., Wu, J., Sloan, S.A., Zhang, Y., Chen, K., Wu, J. Q., Barres, B. A. and Zhang, C., 2015. Systematic discovery of regulated and conserved alternative exons in the mammalian brain reveals NMD modulating chromatin regulators. Proc. natl. Acad. Sci. USA., 112:3445-3450. https://doi.org/10.1073/pnas.1502849112
Zahedi, M., Hojjati, M.R., Fathpour, H., Rabiei, Z., Alibabaei, Z. and Basim, A., 2015. Effect of rheum ribes hydro-alcoholic extract on memory impairments in rat model of Alzheimer’s disease. Iran. J. Pharm. Res. 14: 1197-1206.
Zhang, W., Huo, S-X., Wen, Y-L., Xing, H., Zhang, Q., Li, N., Zhao, D., Sun, X. L., Xu, J., Yan, M. and Chen, X. J., 2015. Pharmacokinetics of acteoside following single dose intragastric and intravenous administrations in dogs. Chinese J. nat. Med., 13: 634-640. https://doi.org/10.1016/s1875-5364(15)30060-1
To share on other social networks, click on any share button. What are these?










