Performance Stability Assessment in upland Cotton Strains throughout Cotton-growing Belt in Pakistan
Research Article
Performance Stability Assessment in upland Cotton Strains throughout Cotton-growing Belt in Pakistan
Muhammad Jamil1*, Ghulam Sarwar2, Imran Akhtar3, Ghayour Ahmad4 and Saeed Ahmad4
1Cotton Research Station Vehari, Pakistan; 2Cotton Research Station, Ayoub Agricultural Research Institute, Faisalabad, Pakistan; 3Regional Agricultural Research Institute, Bahawalpur, Pakistan; 4Cotton Research Institute, old Shujabad Road Multan, Pakistan.
Abstract | Cotton is a real cash crop and contributes to the national economy by enhancing exports. The yield performance of cotton is fluctuating under erratic test conditions due to genotype into environment interaction. The present study was executed at 14sites throughout Pakistan according to a randomized complete block design replicated 03 times during Kharif (summer season) 2019.The experiment aimed to assess the performance stability of studied strains in terms of seed cotton yield against selected environments. Twenty-five upland cotton strains recently bred by different research stations including the check cultivar (CIM-602) were selected for this study. Additive main effects and multiplicative interaction (AMMI) analysis procedure was followed for the data analysis. The analysis of variance revealed that strains, environments and genotype into environments interaction (GEI) results were significant at (p≤0.01). Further, the GEI sum of squares (SS) was comprised of (20.5%) out of total variability. Seven interaction Principal Component Axis (IPCA) were found significant at (p≤0.01). IPCA-1 and IPCA-2 were found enfolded with37.9% and 22.2% of GEI portion respectively. AMMI-1 was diagnosed as a predictive model which delineated all test sites into 4 mega environments. The strain (NIAB-1011) won seven environments and qualified as the overall winner of the trial by giving the highest 2604 Kg ha-1 seed cotton yield. Strain GH-U had yielded a maximum of 4070 Kg ha-1inthe Khuzdar environment. Sahiwal was ranked the top environment with a yield of 3161 Kg ha-1 followed by Khuzdar (2845 Kg ha1) respectively. The genotype selection index (GSI: A non-parametric approach to determine performance stability) distinguished NIAB-135, BH-224 and FH-Am17 being the most stable strains bearing the least GSI value and may be released for general cultivation to boost national cotton production.
Received | February 14, 2022; Accepted | May 25, 2022; Published | October 05, 2022
*Correspondence | Muhammad Jamil, Cotton Research Station Vehari, Pakistan; Email: jamil1091abr@gmail.com
Citation | Jamil, M., G. Sarwar, I. Akhtar, G. Ahmad and S. Ahmad. 2022. Performance stability assessment in upland cotton strains throughout cotton-growing belt in Pakistan. Sarhad Journal of Agriculture, 38(4): 1361-1369.
DOI | https://dx.doi.org/10.17582/journal.sja/2022/38.4.1361.1369
Keywords | Genotype selection index, Mega environments, Pakistan, Performance stability, Upland cotton strains
Copyright: 2022 by the authors. Licensee ResearchersLinks Ltd, England, UK.
This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Introduction
Upland cotton (Gossypium hirsutum. L) is mainly cultivated for its spinnable fibre worldwide. It is truly considered a cash crop and the backbone of the country’s economy. It accounts for 0.8% accumulation in GDP and 4.2 per cent value addition in the agriculture sector (GoP, 2020). Pakistan is numbered as the fifth-best cotton producing and consuming state (Nisar et al., 2022). Due to the increasing demand in the textile sector, it became essential to develop the best yielding cotton cultivars bearing quality lint parameters. Breeding of genotypes possessing genetically improved qualitative and polygenic traits is needed to cope with different types of stresses (Bakhtavar et al., 2015).
In routine, conduction of yield trials constituted with newly bred strains along with check at multi-locations is a general practice in the world. The success of crop breeding schemes depends upon the possibility of releasing cultivars bearing definite superior yields across a set of erratic environmental situations (Hassani et al., 2018). Optimum sowing time is a crucial factor for the best yield in a specific environment. Ishaq et al. (2022) found mid of the March as the best sowing time for upland cotton in the central zone of the Punjab province. Several statistic tools have been used to isolate stable strains against test locations in cotton crop (Phuke et al., 2017). The additive main effects and multiplicative interaction (AMMI) model is an elegant tool to explore the genotype into environments interaction (GEI) segment of variability in multi-location yield experiments (Verma and Singh, 2020). This model is a hybrid of interaction and additive variability segments. This procedure effectively measures additive effects and multiplicative effects at the same time. The principal component analysis (PCA) module is applied to understand signal information present in the GEI segment (Bocianowski et al., 2019). This analysis is highly effective and catches a major part of the variation present in GEI and splitting additive main effects due to genotypes and environments (Ajay et al., 2019). Stable varieties deviate less from the average yield across test locations. In AMMI analysis results will become biased if the first axis of the interaction principal component is squeezed with less portion of interaction variance (Oyekunle et al., 2017). Researchers can change their priority depending upon the aim of the multi-location experiment for a high yield of a variety instead of increased stability in performance (Verma and Singh, 2021).
Yield stability studies in upland cotton strains using AMMI analysis tested at 14 locations including newly emerging cotton pockets of Balochistan and Khyber PakhtunKhwa provinces are missing in the literature. Further, strains tested in this study are newly bred bearing diversified genetic bases never tested earlier. It was postulated that cotton strains with stable yield performance can boost national cotton production. The main purpose of this study was to identify the environment most suitable for the highest cotton yield. Further to quantity stability in yield performance of cotton strains tested over erratic environments and to get approval from concerned authorities for their release.
Materials and Methods
The present experiment was executed at fourteen prominent locations across Pakistan in the National coordinated varietal trial (NCVT) during Kharif (summer season) 2019. The trial was sown at all locations in the first week of May. An experiment was comprised of twenty-five cotton strains (Table 4) bred by different research stations including one check variety CIM-602. The layout of the experiment was a randomized complete block design (RCBD) repeated thrice. The experimental unit was comprised of 5 m long four rows 0.75 m apart from each other. A distance of 0.3 m was maintained between plants after thinning. Irrigation was applied according to the needs of the plant and weather conditions. Fertilizer was applied according to the soil analysis to ensure optimum nutrients available to plants. Recommended agronomic measures were adopted during the growing season. Insect pest populations were kept below economic injury level by spraying recommended agrochemicals. At crop maturity, data of seed cotton yield was collected from all sites repeat wise and converted into kg ha-1.
Data Analysis
Repeat wise data of seed cotton yield in kg ha-1 collected from all sites was analyzed with the analysis of variance (ANOVA) tool (Steel et al., 1997). To measure the segment of GEI for seed cotton yield, data were analyzed to the AMMI procedure described by Gauch (2013). This procedure applies ANOVA to split variability into additive main effects due to genotype, environment and GEI portion. Variability captured by the GEI portion is further analyzed by employing the PCA technique. F-test is used to test the significance of the interaction principal component axis (IPCA) at a given degree of freedom. AMMISOFT version 1.0 available at (https://scs.cals.cornell.edu/people/hugh-gauch) was used for data analysis in the present study. The AMMI equation is as below.
Table 1: Description of 14 test sites along with 25 studied cotton strain.
|
14-Environments |
||||||
|
S.N |
Code |
Description |
Soil Type |
Climate |
||
|
1 |
EN01 |
Central Cotton Research Institute, Multan |
Loam type |
Semi-Arid |
||
|
2 |
EN02 |
4B farm Multan |
Loam type |
Semi-Arid |
||
|
3 |
EN03 |
Cotton Research Station, Bahawalpur |
Sandy loam |
Arid |
||
|
4 |
EN04 |
Cotton Research Station, Sahiwal |
Loam type |
Semi-Arid |
||
|
5 |
EN05 |
Cotton Research Station, Khanpur (Rahim Yar Khan) |
Clay loam |
Arid |
||
|
6 |
EN06 |
Cotton Research Station, Vehari |
Sandy loam |
Semi-Arid |
||
|
7 |
EN07 |
Central Cotton Research Institute, Sakrand |
- |
Arid |
||
|
8 |
EN08 |
Cotton Research Station, Ghotki |
Loamy |
Arid |
||
|
9 |
EN09 |
Cotton Research Station, Mir Pur Khas |
- |
Arid |
||
|
10 |
EN10 |
Nuclear Institute for Agriculture, Tando Jam |
- |
Arid |
||
|
11 |
EN11 |
Cotton Research Station, Lasbella |
Loamy |
- |
||
|
12 |
EN12 |
Cotton Research Station, Sibbi |
- |
|||
|
13 |
EN13 |
Agriculture Research Institute, Khuzdar |
- |
|||
|
14 |
EN14 |
Cotton Research Station, Dera Ismail khan |
Clay |
Temperate |
||
|
25-Strains along with a standard variety |
||||||
|
S.N |
Code |
Description |
S.N |
Code |
Description |
|
|
1 |
GN01 |
NIAB-512 |
15 |
GN15 |
RH-670 |
|
|
2 |
GN02 |
NIAB-973 |
16 |
GN16 |
Himalaya |
|
|
3 |
GN03 |
NIAB-819 |
17 |
GN17 |
GH-sultan |
|
|
4 |
GN04 |
NIAB-135 |
18 |
GN18 |
GH-Uhad |
|
|
5 |
GN05 |
NIAB-1011 |
19 |
GN19 |
FH-Anmol |
|
|
6 |
GN06 |
NIA-89 |
20 |
GN20 |
FH-492 |
|
|
7 |
GN07 |
IUB-73 |
21 |
GN21 |
FH-155 |
|
|
8 |
GN08 |
VH-383 |
22 |
GN22 |
FH-Super 2017 |
|
|
9 |
GN09 |
VH-189 |
23 |
GN23 |
FH-Am 17 |
|
|
10 |
Stnd |
CIM-602 (check) |
24 |
GN24 |
BH-224 |
|
|
11 |
GN11 |
VH-402 |
25 |
GN25 |
BH-223 |
|
|
12 |
GN12 |
SLH-33 |
||||
|
13 |
GN13 |
RH-Kashish |
||||
|
14 |
GN14 |
RH-Afnan-2 |
||||
Yge = µ + αg + βe + Σn λn γgn δen + ρge
Where;
Yge: Yield of genotype g (Kg per ha.) in environment e µ: Grand meanαg Mean Deviation for particular genotype g βe: Mean Deviation from environment means λ n: Singular Value for IPC (Interaction Principal Component) Axis n γgn: represents to Genotype g eigenvector value to IPC axis nδen: the value of eigenvector of environment for IPC axis n ρge: denotes to residual.
Further, AMMI stability value (ASV) was derived to rank cotton strains according to stability parameter by employing the formula given by Purchase (1997) as under:
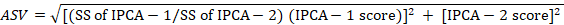
Where;
SS: Sum of Squares. IPCA-1: Interaction Principal Component Axis 1, IPCA-2: Interaction Principal Component Axis 2. As per protocol, lower (ASV) indicates stable genotypes and vice versa.
The genotype selection index (GSI) given by (Farshadfar et al., 2008) was calculated by the following formula. YSI = RASV + RY where RASV: Rank of AMMI Stability Value; RY: Rank of the mean yield across environments.
Table 2: Analysis of variance for seed cotton yield in 25 strains across 14 locations during 2019-20.
|
SOV |
DF |
SS |
MSS |
The proportion of variance % |
||
|
TV |
A&I V |
GEI |
||||
|
Treatments |
349 |
557257931 |
1596728A |
98.3 |
||
|
Strains |
24 |
60161677 |
2506737A |
10.6 |
||
|
Environments |
13 |
380925564 |
29301966A |
67.2 |
||
|
S x E |
312 |
116170690 (Total) 112060101 (Signal) 4110588 (Noise) |
372342A |
20.5 |
||
|
IPCA-1 |
36 |
43981556 |
1221710A |
37.9 |
||
|
IPCA-2 |
34 |
25784853 |
758378A |
22.2 |
||
|
IPCA-3 |
32 |
11965735 |
373929A |
10.3 |
||
|
IPCA-4 |
30 |
9103409 |
303447A |
7.8 |
||
|
IPCA-5 |
28 |
5795883 |
206996A |
5.0 |
||
|
IPCA-6 |
26 |
5248821 |
201878A |
4.5 |
||
|
IPCA-7 |
24 |
4076585 |
169858A |
3.5 |
||
|
IPCA-Residual |
102 |
10213848 |
100136 |
8.8 |
||
|
Error |
700 |
9647297 |
13782 |
1.7 |
||
|
Blocks x Env. |
28 |
793723 |
28347B |
0.1 |
||
|
Pure Error |
672 |
8853575 |
13175 |
1.6 |
||
|
Total |
1049 |
566905228 |
540424 |
100 |
100 |
100 |
A Significant at (p≤0.01)B Significant at (p≤0.05)
Note: F-test uses pure error because blocks x environments are significant at (p≤0.05)
SOV: Source of variance DF: Degree of freedom SS: Sum of squares MSS: Mean sum of squares TV: Total variance A&IV: Additive and Interaction variance GEI: Genotype x Environment interaction IPCA: Interaction Principal Component Axis.
Results and Discussion
ANOVA results related to 25cotton strains tested at 14locations in the country for seed cotton yield are presented in Table 2. The main effects due to stains, environments and GEI were found significant (p≤0.01).This significant GEI segment of variation provided sufficient grounds for AMMI analysis. Environmental main effects were found biggest portion (67.2%) of total variability followed by the GEI segment, which was (20.5%) of total variation present in the studied cotton strains. The main effects of strains were found (10.6%), which is approximately half than GEI effects. These findings are in line with the results given by Riaz et al. (2013), this researcher also found a similar proportion of variability in cotton. The occurrence of significant GEI effects is challenging for breeders in breeding superior cultivars. A successful variety must perform well throughout the areas for which, released for cultivation. The high GEI variation component creates complexity in the assessment of genotype inbuilt genetic potential. It was established fact that yield was deviated by environment main effects and GEI segment (Ntawuruhunga et al., 2001). GEI portion was further analyzed and found that it was composed of more than 96% of signal information. Seven interaction principal components axis (IPCA) were found significant at (p≤0.01). The first two IPCA captured more than (60%) of the GEI portion of variability. The residual portion of IPCA was (8.8%) of GEI. These results are confirmatory to the findings of Krishnamurthy et al. (2021). All seven calculated IPCA captured 91.2% of variability present in the GEI potion.
AMMI Model diagnosis and winner genotypes
AMMI consists of model family members such as AMMI-0, AMMI-1, and AMMI-2 so on possessing 0, 1 and 2 IPCA respectively. Predictive based accuracy, biometrical significance and results interpretability are the top criteria basis for model identification. AMMI-0 represents a simple linear model without any interaction segment of variability. GEI portion left behind capturing by last IPCA was treated as residual. Early IPCA usually picks the signal information portion (a portion of data from which we can draw some conclusion) while the last few IPCA and residual capture mostly noise. In the present study (96.5%) GEI was measured as a signal portion and leftover 3.5% as noise. AMMI-F denoted a full model consisting of all GEI segments and no residual portion. AMMI-F was considered near to raw data and lacks any practical utilization.
As argued by Gauch (2013), in a well-known published article relevant to AMMI analysis, the model diagnosis was not executed. Instead, AMMI-1 was chosen as the default model because appropriate bi-dimensional graphs are possible to plot in such models. For simplicity during mega environment delineation AMMI-1 model was also selected by Agahi et al. (2020). A however simpler model like AMMI-1 would be more suitable due to practical simplicity that involves a few mega-environments, which is justified when the most emphasis was put on the usage of wider adaptation. Genotype NIAB-1011 was found winner of the yield trial and won all AMMI model family (Table 3). According to theAMMI-1 default model, NIAB-1011 won a maximum of seven environments followed by GH-U and NIAB-135 with the winner of three environments each. VH-189 also won one environment in the AMMI-1 model.
Table 3: Winners of AMMI model family across 14 locations during 2019-20.
|
Strains |
AMMI model family |
||||||||
|
0 |
1 |
2 |
3 |
4 |
5 |
6 |
7 |
F |
|
|
GH-Uhad |
3 |
2 |
3 |
3 |
2 |
3 |
3 |
3 |
|
|
NIAB-1011 |
14 |
7 |
8 |
5 |
5 |
6 |
5 |
5 |
3 |
|
GH-Himaliya |
1 |
1 |
1 |
2 |
|||||
|
FH-Super 2017 |
1 |
1 |
1 |
||||||
|
GH-Sultan |
1 |
1 |
|||||||
|
RH-670 |
1 |
1 |
1 |
1 |
1 |
1 |
|||
|
NIAB-135 |
3 |
2 |
1 |
2 |
2 |
2 |
1 |
1 |
|
|
NIA-89 |
1 |
2 |
1 |
1 |
1 |
||||
|
VH-189 |
1 |
1 |
2 |
2 |
1 |
1 |
1 |
1 |
|
|
Mega environments |
1 |
4 |
5 |
6 |
6 |
7 |
7 |
8 |
9 |
AMMI: Additive Main effects and Multiplicative Interaction.
Delineation of mega environments
A ranking of the five best yielding cotton strains through 14 testing environments in AMMI-1 and AMMI-F model results are presented in Table 4. Test environments are arranged in the table according to IPCA-1 scores so that upper and bottom-placed sites bear opposite GEI interaction behavior. Test sites and genotypes are enlisted in Table 1 along with codes used in biplots. In the AMMI-1 model 14 environments were delineated into 4 mega environments (ME). A mega environment is a combination of test sites with statistically non-significant effects on yield. ME-1 was the largest and won by GN05 (NIAB-1011) consisting of 7 environments DI Khan, Ghotki and all Punjab locations except Sahiwal.ME-2 won by strain GN18 (GH-Uhad) across locations of Sakrand, Lasbella and Khuzdar (Figure 1). ME-3 consists of three sites Viz: Sahiwal, Mir Pur Khas & Tando Jam in Sindh province and won by GN04 coded for strain NIAB-135. ME-4 was the smallest and won by GN09 (VH-189) against a single site of Sibbi, Balochistan. Signal information in the AMMI-F model is complicated, complex and difficult to interpret. The ratio mentioned in (Table 4) is obtained as a ratio of winner genotype in a particular environment divided by the yield of the overall winner, which is GN05 (NIAB-1011) in the present case (Gauch, 2008). So the strain GN18 (GH-Uhad) bears a yield advantage of 29.4% across site EN11 (Lasbella) and EN13 (Khuzdar) 24.5% in enhanced yield than the overall winner genotype. This edge was due to minor adaptations acquired by respective strains at specific sites due to GEI interaction. Similarly, GN04 (NIAB-135) possessed a yield advantage of (8.4-11.3%) across sites Sahiwal, Mir Pur Khas & Tando Jam respectively (Table 4).
Identification of stable cum yielder cotton strains
A lot of cotton varieties were released in the country in the past but most of them were not survived in the field except NIAB-78 and MNH-93 after 5 years of their release due to unstable yield across changing environments. Riaz et al. (2013) also found uneven behavior of cotton genotypes for stability regarding the seed cotton yield. Breeding for adaptable varieties
Table 4: Ranking of top 5 cotton strains according to AMMI-1 and AMMI-F model families for 25 cotton strains in 4 mega environments.
|
Mega Env. |
Env. code |
Ratio |
AMMI-1 ranks |
AMMI-F ranks |
||||||||
|
1 |
2 |
3 |
4 |
5 |
1 |
2 |
3 |
4 |
5 |
|||
|
ME-2 |
EN11 |
1.0294 |
GN18 |
GN05 |
GN16 |
GN17 |
GN22 |
GN18 |
GN05 |
GN16 |
GN17 |
GN22 |
|
EN13 |
1.0245 |
GN18 |
GN05 |
GN16 |
GN17 |
GN22 |
GN18 |
GN05 |
GN16 |
GN22 |
GN17 |
|
|
EN07 |
1.0059 |
GN18 |
GN05 |
GN16 |
GN17 |
GN22 |
GN05 |
GN16 |
GN22 |
GN18 |
GN17 |
|
|
ME-1 |
EN02 |
1 |
GN05 |
GN18 |
GN16 |
GN17 |
GN22 |
GN05 |
GN18 |
GN04 |
GN17 |
GN19 |
|
EN08 |
1 |
GN05 |
GN18 |
GN16 |
GN17 |
GN22 |
GN17 |
GN05 |
GN21 |
GN18 |
GN22 |
|
|
EN05 |
1 |
GN05 |
GN18 |
GN16 |
GN17 |
GN22 |
GN05 |
GN04 |
GN25 |
GN18 |
GN01 |
|
|
EN01 |
1 |
GN05 |
GN18 |
GN16 |
GN17 |
GN04 |
GN18 |
GN04 |
GN16 |
GN05 |
GN17 |
|
|
EN06 |
1 |
GN05 |
GN18 |
GN16 |
GN04 |
GN17 |
GN16 |
GN04 |
GN01 |
GN18 |
GN22 |
|
|
EN03 |
1 |
GN05 |
GN04 |
GN18 |
GN16 |
GN17 |
GN22 |
GN13 |
GN11 |
GN04 |
GN01 |
|
|
EN14 |
1 |
GN05 |
GN04 |
GN18 |
GN16 |
GN17 |
GN16 |
GN17 |
GN18 |
GN11 |
GN09 |
|
|
ME-3 |
EN04 |
1.0084 |
GN04 |
GN05 |
GN18 |
GN16 |
GN09 |
GN09 |
GN01 |
GN08 |
GN15 |
GN18 |
|
EN09 |
1.0226 |
GN04 |
GN05 |
GN09 |
GN25 |
GN18 |
GN04 |
GN21 |
GN05 |
GN25 |
GN17 |
|
|
EN10 |
1.1130 |
GN04 |
GN09 |
GN25 |
GN01 |
GN15 |
GN15 |
GN06 |
GN03 |
GN04 |
GN16 |
|
|
ME-4 |
EN12 |
1.2969 |
GN09 |
GN04 |
GN25 |
GN02 |
GN03 |
GN06 |
GN08 |
GN04 |
GN25 |
GN02 |
AMMI: Additive Main effects and Multiplicative Interaction.
across a wide range of target locations is the real challenge to the breeders (Bose et al., 2014). AMMI stability value (ASV) was calculated for cotton strains studied based on IPCA-1 and IPCA-2 scores (Table 5). Bigger the absolute value of ASV, the better the adaptability of a particular genotype for a certain location. On the contrary, smaller ASV values highlight genotype general stability across tested environments. Stability itself is not a reliable selection indicator as stable cultivars were mostly found to be poor yielders (Mohammadi et al., 2017), so the use of yield plus stability as a single non-parametric index is generally required (Farshadfar et al., 2008). Genotype selection index (GSI) was obtained by adding ranks of ASV and yield of the respective strain at that particular site. This index depicted GN04 (NIAB-135) and GN24 (BH-224) followed by GN23 (FH-Am 17) and GN14 (RH-Afnan-2) as stable strains bearing minimum GSI values respectively. (Table 5). GN04 (NIAB-135) bears little edge of high yield, while GN24 (BH-224) was more stable than the former strain. On the other hand, GN06 (NIA-89) followed by GN02 (NIAB-973) were proved as poor yielders and unstable in performance across test sites respectively. AMMI-2 biplot indicated strains plotted near origin represent stable types while strains on the periphery were unstable entries in this trial (Figure 2). Similarly, stable and high yielder group was encircled separately from poor yielder and unstable cotton strains.
Conclusions and Recommendations
The present experiment highlighted that cotton strains behaved differently to test environments. Strain NIAB-1011 was found optimum yielder and suitable for DI Khan, Ghotki and all Punjab locations except the Sahiwal site.GH-U was suitable for the new emerging sites of Balochistan and bears a 24-30% yield advantage due to minor adaptations. Three
Table 5: Ranking of 25 strains of cotton for mean yield (Kgha-1), AMMI stability value (ASV)& genotype selection index.
|
Strains |
Code |
Mean yield |
Rank |
IPCA-1 score |
IPCA-2 score |
ASV |
Rank |
GSI |
|
NIAB-512 |
GN01 |
2184 |
8 |
-3.059 |
-16.211 |
17.0 |
12 |
20 |
|
NIAB-973 |
GN02 |
1856 |
22 |
-18.080 |
5.767 |
31.4 |
21 |
43 |
|
NIAB-819 |
GN03 |
1898 |
20 |
-13.822 |
8.892 |
25.2 |
17 |
37 |
|
NIAB-135 |
GN04 |
2365 |
5 |
-6.183 |
-9.412 |
14.1 |
7 |
12 |
|
NIAB-1011 |
GN05 |
2604 |
1 |
19.293 |
-7.915 |
33.8 |
22 |
23 |
|
NIA-89 |
GN06 |
1882 |
21 |
-12.379 |
33.240 |
39.4 |
24 |
45 |
|
IUB-73 |
GN07 |
1672 |
25 |
-7.814 |
-9.848 |
16.6 |
11 |
36 |
|
VH-383 |
GN08 |
2062 |
15 |
-8.312 |
-2.292 |
14.4 |
8 |
23 |
|
VH-189 |
GN09 |
2022 |
16 |
-19.796 |
-9.531 |
35.1 |
23 |
39 |
|
CIM-602(check) |
Stnd |
1954 |
18 |
6.491 |
1.163 |
11.1 |
3 |
21 |
|
VH-402 |
GN11 |
1843 |
23 |
-8.781 |
0.050 |
15.0 |
9 |
32 |
|
SLH-33 |
GN12 |
1920 |
19 |
-6.668 |
-1.830 |
11.5 |
5 |
24 |
|
RH-Kashish |
GN13 |
1691 |
24 |
1.324 |
0.273 |
2.3 |
1 |
25 |
|
RH-Afnan-2 |
GN14 |
2084 |
13 |
6.331 |
-4.898 |
11.9 |
6 |
19 |
|
RH-670 |
GN15 |
2201 |
7 |
-1.634 |
21.173 |
21.4 |
15 |
22 |
|
GH-Himalaya |
GN16 |
2434 |
3 |
18.022 |
3.890 |
31.0 |
20 |
23 |
|
GH-sultan |
GN17 |
2408 |
4 |
17.216 |
0.381 |
29.4 |
18 |
22 |
|
GH-Uhad |
GN18 |
2531 |
2 |
25.602 |
2.428 |
43.7 |
25 |
27 |
|
FH-Anmol |
GN19 |
2071 |
14 |
11.937 |
-1.020 |
20.4 |
13 |
27 |
|
FH-492 |
GN20 |
1995 |
17 |
-10.835 |
-15.564 |
24.2 |
16 |
33 |
|
FH-155 |
GN21 |
2108 |
11 |
-0.657 |
-15.227 |
15.3 |
10 |
21 |
|
FH-Super 2017 |
GN22 |
2327 |
6 |
17.226 |
4.096 |
29.7 |
19 |
25 |
|
FH-Am 17 |
GN23 |
2174 |
9 |
2.481 |
10.508 |
11.3 |
4 |
13 |
|
BH-224 |
GN24 |
2164 |
10 |
4.311 |
2.104 |
7.6 |
2 |
12 |
|
BH-223 |
GN25 |
2098 |
12 |
-12.216 |
2.207 |
21.0 |
14 |
26 |
cotton strainsNIAB-135, BH-224 and FH-Am 17 were found to yield cum stable types. Their release from respective seed councils for general cultivation may be perused to boost cotton production in the country.
Acknowledgements
The authors of this manuscript are thankful to the Pakistan central cotton committee (PCCC) team for providing a platform and coordination in this study.
Novelty Statement
Genotype into environment interaction (GEI) study in cotton by AMMI method at 14 diversified locations including new emerging cotton sites of Khyber Pakhtunkhwa and Balochistan is rarely explored in Pakistan.
Author’s Contribution
Muhammad Jamil: Conducted the trial at the Vehari site and wrote the manuscript.
Ghulam Sarwar: Critically read the manuscript and added input.
Imran Akhtar: Prepared figures and tables.
Ghayour Ahmad: Analyzed the data with software.
Saeed Ahmad: Reviewed recent literature.
Conflict of interest
The authors had declared no conflict of interests regarding this article.
References
Agahi, K., J. Ahmadi, H.A. Oghan, M.H. Fotokian and S.F. Orang. 2020. Analysis of genotype into environment interaction for seed yield in spring oilseed rape using the AMMI model. Crop Br. Appl. Biotechnol., 20(1): e26502012,2020. https://doi.org/10.1590/1984-70332020v20n1a2
Ajay, B.C., J. Aravind, A.R. Fiyaz, K. Narendra, L. Chuni, K. Gangadhar, K. Praveen, M.C. Dagla and S.K. Bera. 2019. Rectification of modified AMMI stability value (MASV) Indian J. Genet., 79(4):726- 731. https://doi.org/10.31742/IJGPB.79.4.11
Bakhtavar, M.A., I. Afzal, S.M.A. Basra, A.U.H. Ahmad and M.A. Noor. 2015. Physiological strategies to improve the performance of Spring Maize (Zea mays L.) planted under early and optimum sowing conditions. PLoS One, 10(4): 1-15. https://doi.org/10.1371/journal.pone.0124441
Bocianowski, J., J. Niemann and K. Nowosad. 2019. Genotype-by environment interaction for seed quality traits in interspecific cross-derived Brassica lines using additive main effects and multiplicative interaction model. Euphytica, 215(7):1–13. https://doi.org/10.1007/s10681-018-2328-7
Bose, L.K., N.N. Jambhulkar, K. Pande and O.N. Singh. 2014. Use of AMMI and other stability statistics in the simultaneous selection of rice genotypes for yield and stability under direct-seeded conditions. Chillian J. Agric. Res., 7(1): 1-9.
Farshadfar, E. 2008. Incorporation of AMMI stability value and grain yield in a single non-parametric index (GSI) in bread wheat. Pak. J. Biol. Sci., 11:1791–1796. https://doi.org/10.3923/pjbs.2008.1791.1796
Gauch, H.G., H.P. Piepho and P. Annicchiarico. 2008. Statistical analysis of yield trials by AMMI and GGE: further considerations. Crop Sci., 48:866–889. https://doi.org/10.2135/cropsci2007.09.0513
Gauch, H.G. 2013.A Simple Protocol for AMMI Analysis of Yield Trials. Crop Sci., 53: 1860-1869. https://doi.org/10.2135/cropsci2013.04.0241
GoP. 2020. Agricultural Statistics Year Book 2019-20. Economic Survey, Finance Division, Economic Advisor’s Wing, Govt. of Pakistan, Islamabad.
Hassani, M., B. Heidari, A. Dadkhodaie and P. Stevanato. 2018. Genotype by environment interaction components underlying variations in root, sugar and white sugar yield in sugar beet (Beta vulgaris L.). Euphytica, 214(79):1-21 https://doi.org/10.1007/s10681-018-2160-0
Ishaq, M.Z., U. Farooq, M.A. Bhutta, S. Ahmad, A. Bibi, H.U. Rehman, U. Farooq, J. Ashraf and S. Nisar. 2022. Effect of Sowing dates and genotypes on Yield and Yield Contributing traits of Upland Cotton (Gossypium hirsutum L.). Sarhad J. Agric., 38(1): 16-25. https://doi.org/10.17582/journal.sja/2022/38.1.16.25
Krishnamurthy, S.L. and P.C., Sharma. 2021. Additive main effects and multiplicative interaction analyses of yield performance in rice genotypes for general and specific adaptation to salt stress in locations in India. Euphytica., 217(20):1-15. https://doi.org/10.1007/s10681-020-02730-7
Mohammadi, R., M. Armion, A. Shabani and A. Daryaei.2017. Identification of stability and adaptability in advanced durum wheat genotypes using AMMI analysis. Asian J. Plant Sci., 6(42):1261-1268. https://doi.org/10.3923/ajps.2007.1261.1268
Nisar, N., T.M. Khan, M.A. Iqbal, R. Ullah, M.A. Bhutta, S. Ahmad, A. Bibi, H.U. Rehman, U. Farooq and M.Z. Ishaq. 2022. Assessment of Yield Contributing Quantitative Traits in Upland Cotton (Gossypium hirsutum). Sarhad J. Agric., 38(1): 353-359. https://doi.org/10.17582/journal.sja/2022/38.1.353.359
Ntawuruhunga, P.H., P.R. Rubaihayo, J.B.A. Whyte, A.G.O. Dixon and D.S.O. Osiru. 2001. Additive main effects and multiplicative interaction analysis for storage root yield of cassava genotypes evaluation in Uganda. Afr. Crop. Sci. J., 9: 591-598. https://doi.org/10.4314/acsj.v9i4.27581
Oyekunle, M., A. Menkir, H. Mani, G. Olaoye, I.S. Usman and S.G. Ado. 2017. Stability analysis of maize cultivars adapted to tropical environments using AMMI analysis. Cereal Res. Commun., 45: 336–345. https://doi.org/10.1556/0806.44.2016.054
Phuke, R.M., K. Anuradha, K. Radhika, F. Jabeen, G. Anuradha, T. Phuketsh. 2017. Genetic variability, genotype x environment interaction, correlation, and GGE biplot analysis for grain iron and zinc concentration and other agronomic traits in RIL population of sorghum (Sorghum bicolor L. Moench). Front Plant Sci., 8(12): 22-29. https://doi.org/10.3389/fpls.2017.00712
Purchase, J.L.1997. Parametric analysis to describe G x E interaction and yield stability in winter wheat. PhD Thesis. Department of Agronomy, Faculty of Agriculture, University of the Orange Free State, Bloemfontein, South Africa.
Riaz, M., M. Naveed, J. Farooq, A. Farooq, A. Mahmood, C. Rafiq, M. Nadeem and A. Sadiq. 2013. AMMI analysis for stability, adaptability and GE interaction studies in cotton (Gossypium hirsutum L.). J. Anim. Plant Sci., 23(3):865-871.
Steel, R.G.D., J.H. Torrie and D.A. Dickey. 1997. Principles and procedure of statistics: A biometrical approach 3rd ed. McGraw-Hill Book Co., New York.
Verma, A. and G.P. Singh. 2020. Gx E Interaction Analysis of wheat genotypes evaluated under restricted irrigated timely sown conditions of north eastern plains zone using AMMI & yield stability measures. Int. J. Curr. Microbiol. App. Sci., 9(11): 957-970. https://doi.org/10.20546/ijcmas.2020.911.114
Verma, A. and G.P. Singh. 2021. AMMI with BLUP analysis for stability assessment of wheat genotypes under multi-locations timely sown trials in the Central Zone of India. J Agric. Sci. Food Technol., 7(1): 118-124. https://doi.org/10.17352/2455-815X.000098
To share on other social networks, click on any share button. What are these?








