Isolation and Identification of Newcastle Disease Viruses from Naturally Infected Chickens
Special Issue:
Emerging and Re-Emerging Animal Health Challenges in Low and Middle-Income Countries
Isolation and Identification of Newcastle Disease Viruses from Naturally Infected Chickens
Yasser Asaad Hameed Al-Shareef, Firas Hussain Kadim Abawi*
Pathology and Poultry Disease Department, College of Veterinary Medicine, Al-Qasim Green University, Babylon 51013, Iraq.
Abstract | Newcastle disease (ND) is one of the most infectious and lethal infections in poultry attributed by severe economic losses and undermining poultry well-being. The ND can cause high mortalities in all types of poultry nevertheless of vaccination status, indicating breakthrough infections. In order to investigate the nature and dynamics of these breakthrough infections, samples were collected from infected flocks with existing clinical signs of the disease. Attempts to isolate, identify and characterize positive samples were made by inoculating chicken embryonated eggs followed by haemagglutination and haemagglutination inhibition assays. Using reverse transcriptase-polymerase chain reaction, the presence of the virus was confirmed and mean death time approach was applied to determine the pathotype of isolates (velogenic). Next, we assessed the most important determinant of ND virus pathogenicity by the analysis of the fusion protein cleavage sites, phylogenetic analysis sand compared genomics of NDV isolates to publish sequences. Isolates characterized here showed 90.37% sequence similarity to the Iranian isolate Asil/IR/AAA158/2019 (MN370894.1) within velogenic clusters. Taken together, clinical, genetics and molecular biological approaches identified velogenic strains of NDV in poultry which breach the immunity induced by the currently applied vaccines, warranting future monitoring to better device control strategies in poultry.
Keywords | Newcastle disease, Fusion protein, Phylogenetic tree, Sequencing reverse transcriptase, Polymerase chain reaction, NCBI, MDT, ECE, Broilers
Received | August 01, 2024; Accepted | October 02, 2024; Published | October 25, 2024
*Correspondence | Firas Hussain Kadim Abawi, Pathology and Poultry Disease Department, College of Veterinary Medicine, Al-Qasim Green University, Babylon 51013, Iraq; Email: yah.alshareef1@gmail.com
Citation | Al-Shareef YAH, Abawi FHK (2024). Isolation and identification of Newcastle disease viruses from naturally infected chickens. J. Anim. Health Prod. 12(s1): 32-39.
DOI | http://dx.doi.org/10.17582/journal.jahp/2024/12.s1.32.39
ISSN (Online) | 2308-2801

Copyright: 2024 by the authors. Licensee ResearchersLinks Ltd, England, UK.
This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Introduction
Newcastle disease virus (NDV) is one of the major poultry pathogens that seriously endangers the poultry industry, resulting in highly contagious septic, a fatal and destructive disease affecting a wide variety of poultry and wild birds worldwide. The NDV belongs to the genus Avian orthoavulavirus 1 of the family Paramyxoviridae (Amarasinghe et al., 2018; Dimitrov et al., 2019; Ewies et al., 2017). NDV is an enveloped virus with a non-segmented, single-stranded RNA genome of negative polarity of approximately 15.2 kb (Mayo, 2002). The ND is endemic in various developing countries, including Iraq and causes high economic impact, due to the lethality of the disease. As a result, the poultry industry is facing losses of millions of dollars worldwide (Susta et al., 2011).
The NDV isolates are grouped into five serotypes depending on their severity which is determined by in vivo pathogenicity test parameters, including the mean death time in chicken embryos and the intra-cerebral pathogenicity index in day-old chicks (Alexander, 2000). Based on clinical signs, these are asymptomatic and with invisible intestinal infection that are associated with subclinical signs. The lentogenic strains are characterized by unapparent or subclinical, and mild respiratory signs, with little drop in egg production, nervous signs and negligible mortality. Mesogenic strains are characterized with rare respiratory and nervous signs, if present it appeared as mild signs with coughing (Huang et al., 2004). Young birds are more susceptible in comparison to adult and might show considerable mortality rates (up to 10%), loss of body weight, late nervous signs and decrease in egg production in layers might be observed (OIE, 2012). Neurotropic velogenic disease can appear suddenly, with acute respiratory signs, un-appetence, nervous signs and drop in egg production of layer and viscerotropic velogenic are characterized by severe intestinal haemorrhagic lesions, clear depression, and dehydration, the birds collapsed due to severe greenish yellow diarrhea (OIE, 2012). Drop in egg production in layers, increased respiration and high mortality rate (up to 90%) can be observed. Some birds die suddenly or show nervous signs especially those survived the infection (Terregino and Capua, 2009). It has been shown that additives of Zingiber to broiler chicks feed results in increases in hemagglutination inhibition (HI) titers against ND virus (Al-Bawi and Rabee, 2020), however additives of nano α-tocopherol acetate plus selenium with diet lead to increase antibody titers against ND (Abdulameer et al., 2019).
This study aims to characterize NDV isolates from vaccinated flock which are showing clinical disease. Molecular characterization of NDVs facilitated to evaluate the virulence nature of isolates by sequence analysis of amino acid of the cleavage site of fusion (F) protein. The findings provide a broader understanding of the NDVs and identify a wild-type NDV from infected broiler farms of Babylon province.
Materials and Methods
Trachea and cloacal swabs were collected from live chickens, while spleen, lung, heart, liver, trachea, intestines, brain and kidneys were carried out from infected flocks. According to the OIE manual procedure, all swabs’ samples were treated separately (OIE, 2004) by using isotonic phosphate buffered saline (PBS) (pH 7.0 –7.4) to preserve samples of other organs, until processing. All samples were transported aseptically in cold box chilled directly to the Virology Lab at Veterinary Hospital in Babylon province, and were frozen at –86 °C until examination.
Isolation of the virus was performed in embryonated hen eggs at 10 days old embryo, following OIE manual. For this purpose, 20 eggs were used where 15 eggs were inoculated with isolates and 5 eggs were kept as control. Stock processed sample was inoculated in allantoic cavity using 0.1 ml of processed sample for each egg. Control embryonated chicken eggs were inoculated by the same method except sterile normal saline was used. All opened shells were closed by wax and the eggs were incubated at 37ºC and observed daily. Any death of embryos 24 hours post infection was excluded and regarded as non-specific death. The allantoic fluid (AF) was collected and stored according to the routine and standard procedure proposed by the OIE manual.
According to previous studies, the presence of NDV in AF can be determined by real-time RT-PCR, haemagglutination (HA) and haemagglutination inhibition (HI) tests (Al-Bawi and Hussein, 2020; Eid et al., 2022). Tissue homogenates of post-mortem samples, clinical samples, and AF were subjected for slide as well as micro-plate HA test to determine the presence of haemagglutinating virus using 1.5% and 0.5% freshly prepared chicken RBC suspension (Stephen et al., 1975). The HI test using anti-NDV hyperimmune sera raised in chickens was employed with the HA positive samples for identification of NDV.
Pathotyping determination via MDT
Mean death time (MDT)
Tenfold (10-6 to 10-9) dilutions of fresh infective AF in sterile phosphate-buffered saline (PBS) were prepared from each dilution and 0.1 mL was inoculated into the allantoic cavities of five 10 days old embryonated chicken eggs another five eggs inoculated with 0.1ml of PBS. The inoculated eggs were incubated at 37°C, examined twice daily for 7 days and the times of the embryo deaths were recorded. The MDT has been used to characterize the NDV pathogenicity as follows: Velogenic, less than 60 hrs; mesogenic, 60 to 90 hrs; and lentogenic, more than 90 hrs (Alexander, 2000).
Titration of NDV isolates
The embryo lethal dose 50 (ELD50) of each isolate was determined using the method of Reed and Muench (1938). To determine the ELD50, 10-fold serial dilutions of AF sample were used at 10-1, 10-2, 10-3, 10-4, 10-5 and 10-6 dilutions. Each of these dilutions was inoculated into 5 embryonated chicken eggs. Mortality was recorded up to five days post inculcation, The dilution of inoculum producing 50 percent dead of embryonated egg was determined. The Reed Muench formula was used to calculate the ELD50:
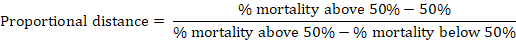
The index was calculated by applying this equation to the dilution that produced lethal rate directly more than 50%.
Molecular identification of NDV
PCR master mix preparation
GoTaq® G2 Green Master Mix is a ready-to-use mixture of high-quality Taq DNA Polymerase, deoxynucleotides, and reaction buffer in a 2X concentration (Table 1). It contains all the necessary reagents for amplification of cDNA. The GoTaq® G2 Green Master Mix contains an inert green dye and a stabiliser which allow direct loading of the final products onto a gel for analysis done according to company instructions as following. The primer used in the PCR amplification are listed in Table 2.
Table 1: Preparation of PCR solutions.
| Components | Concentration | Volume (50 µl) |
| GoTaq® G2 green master mix | 1X | 25 µl |
| Forward primer | 10 µM/µl | 4 µl |
| Reverse primer | 10 µM/µl | 4 µl |
|
ddH2O |
- | 13 µl |
| DNA | 40 ng | 4 µl |
PCR thermocycler conditions
PCR thermocycler conditions were performed by using conventical PCR thermo-cycler system for all genes as following:
| Phase | Tm (ᵒC) | Time | Cycles |
| Initial denaturation |
94 oC |
5 min | 1X |
| Denaturation |
94 oC |
30 sec. | 35X |
| Annealing |
50-58 oC |
30 sec. | |
| Extension |
72 oC |
1 min | |
| Final extension |
72 oC |
5 min | 1X |
Results and Discussion
NDV isolates from pre-vaccinated chickens
A total of 5 flocks in Babylon province (Y03, Y6), Al-Hashimiyah area (Y11, Y12), in Al-Kifil area, (Y13) and in Al-Muhaweel area were reported to exhibit ND clinical signs with a mortality rate ranged from 25-90% (death occurred within 24-96 hrs post- onset of clinical signs). The most common clinical signs were greenish diarrhea, incoordination, swelling of eyelids and head, respiratory sign, torticollis and death. The most commonly observed postmortem lesions were marked hemorrhagic ulcers in the intestinal wall and cecal tonsils, pin point hemorrhages at the tip of proventriculus glands, hemorrhagic lungs, tracheitis with congestion and catarrhal exudates (Figure 2A, B, C and Table 1).
ND struck the poultry industry in the Babylon province of Iraq causing severe economic losses. However, many governmental and private poultry farms were established intensively in Iraq in the last two decades. These farms suffered from ND and circulating ND virus. All commercial chicken farms apply common vaccination schedule against ND which is consisting of live LaSota virus administered in different routes at 1 days of age, booster dose at 10-14 days of age and repeated at 20-24 day of age. Inactivated vaccine had been used in some flocks at 1 day of age. Therefore, this study was carried out to isolate and pathotype field viruses among chickens and characterizes these isolates according to their virulence in this area. The most commonly observed postmortem lesions were marked hemorrhagic ulcers in the intestinal wall and cecal tonsils (Figure 2A), pin point hemorrhages at the tip of proventriculus glands (Figure 2B), hemorrhagic lungs, tracheitis with congestion and catarrhal exudates (Figure 2C, Table 1) as are commonly observed previously (Pazhanivel et al., 2002).
Virus identification
Fifteen NDV suspected field samples were pooled according to their sub-districts in 5 tubes. These tubes were homogenized and suspended for confirmation of ND virus via PCR. The sample positive in PCR were propagated in 10 days old embryonated chicken egg (ECE) via allantoic sac. In all positive cases, embryos died within 24 to 96 hrs post inoculation, where 5 showed positive rapid HA within few seconds indicating that the isolates were hemagglutinating viruses. All HA positive embryo died within 40 to 96 hrs post-inoculation. The rapid HA positive samples were titrated using micro-HA test and the titers ranged from 1:32 - 1:512 (Table 2).
Three positive AF samples have inhibited anti-NDV hyper-immune serum using HI test except two which potentially indicate breakthrough NDV infection (Table 2). The HI titers of the positive viruses ranged from 1:16 - 1:256 (Table 2), as reported earlier (Spackman et al., 2003; Manin et al., 2002).
Table 2: Primers used in this study.
| Organism | Gene | Primer name | 5'-3' | Size (bp) | Accession number | Reference |
| NDV | F | F | ATGGGC(C/T)CCAGA(C/T)CTTCTAC | 535 | KF494201.1 |
Liang et al., 2002 |
| R | CTGCCACTGCTAGTTGTGATAATCC |
PCR product of all the 3 samples revealed a band of supposed size of 535 bp that confirmed the presence of NDV (Figure 1). These isolates (i.e., YS03, YS11, YS12) produced highest mortalities with a rate of 80%, 75% and 50%, respectively and less than 60hr MDT indicating they were velogenic NDV (Tables 1, 3), in agreement with finding Al-Zuhariy et al. (2017).
ND is recognized as one of the foremost threats and causes serious economic losses in the poultry industry despite the intensive vaccination regimes applied against ND. In Iraq, ND is still recognized to be endemic and conventional diagnosis of ND via virus isolation on embryonated eggs followed by serological identification in hemagglutination inhibition test is laborious and time consuming. The speed of the diagnosis has been increased by using methods based on molecular biology such as Reverse transcriptase- CR (Aldous and Alexander, 2001; Jestin and Jestin, 1991). Reverse transcriptase- PCR for the detection of NDV was first described by Jestin and Jestin (1991), Ahmed and Odisho (2018) and to date it has been successfully developed in different modifications (Aldous and Alexander, 2001) using universal primers to detect all NDVs (Jestin and Jestin, 1991). Additionally, Śmietanka et al. (2006) have applied RT-PCR method for the detection and identification of NDV in allantoic fluids of infected embryonated eggs (Alexander, 2008). In this study, real time RT-PCR method was applied for the detection and identification of NDV in pooled NDV samples in three area at Babylon province of Iraq Al-Hashimiyah, Al-Kifil and Al-Muhaweel. Positive samples inoculated in to 10 days old embryonated egg, alantoic fluid gave positive reaction in HA test. The hemagglutination activity was inhibited by NDV hyper immune serum using HI assay to confirm NDV as applied by Zhao et al. (2001). These results correspond to the finding of Spackman et al., showing real time RT-PCR correlated with virus isolation and is significantly faster but cannot detect nucleic acid in all samples (Spackman et al., 2003).
Phylogenetic analysis
The phylogenetic analysis was carried out for three isolate of NDV Iraqi isolates (Chicken/Broiler/babill/2023) (YS03, YS11, YS12) and sequences were submitted to the NCBI and are available under the accession number (PP832924.1, PP832925.1, PP832926.1). The field isolates (YS03, YS11, YS12) showed 99.37% sequence similarity with Iranian isolate Asil/IR/AAA158/2019 (MN370894.1) (Table 4). The F protein based phylogenetic relationships of Iraqi NDV isolated was conducted with other NDV sequences (Figure 4). The evolutionary history was inferred using the Neighbour-Joining method (Saitou and Nei, 1987). The optimal tree with the sum of branch length= 0.01473775 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) are shown next to the branches (Felsenstein, 1985). The tree was drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Jukes-Cantor method (Jukes and Cantor, 1969) and are in the units of the number of base substitutions per site. The analysis involved a total 18 sequences and codon positions included were 1st + 2nd + 3rd + Noncoding. All positions containing gaps and missing data were eliminated. There was a total of 477 positions in the final dataset. Evolutionary analyses were conducted in MEGA7 (Kumar et al., 2016).
Table 1: Investigation data of the NDV among chicken’s flock in (Babylon province) during 2023.
Table 2: Isolation and identification of ND Viruses from naturally infected chickens in (Babylon province) during 2023.
| Location | Sample | Type of chicken | Age (day) | No. passage | (log2) HA | (log2) HI | Real time -PCR | CT value |
|
Babylon Al-Hashimiyah |
YS03 | Broiler | 28 | 3 | 128 | 64 | Positive | 13.928 |
| Babylon Al-Kifil | YS11 | Broiler | 31 | 3 | 32 | 16 | Positive | 24.770 |
| Babylon Al-Kifil | YS12 | Broiler | 35 | 3 | 512 | 256 | Positive | 33.940 |
Table 3: Pathotyping of isolated NDV from naturally infected chickens in (Babylon province) during 2023.
| Location | Sample | Type of chicken | Age (day) | No. passage | ELD50 | MDT (hrs) | Pathotype |
| Babylon Al-Hashimiyah | YS03 | Broiler | 28 | 3 | -8.910 | 58 | Velogenic |
| Babylon Al-Kifil | YS11 | Broiler | 31 | 3 | -8.990 | 57 | Velogenic |
| Babylon Al-Kifil | YS12 | Broiler | 35 | 3 | -8.839 | 59 | Velogenic |
Table 4: Identity of isolates with previously isolated strains of Newcastle virus.
| Isolate | Sequence cover | E value | Identity % | Accession number |
| Avian paramyxovirus 1 isolate YS12 fusion protein (F) gene | 100% | 0 | 100 | PP832926.1 |
| Avian paramyxovirus 1 isolate YS11 fusion protein (F) gene | 100% | 0 | 100 | PP832925.1 |
| Avian paramyxovirus 1 isolate YS03 fusion protein (F) gene | 100% | 0 | 100 | PP832924.1 |
| Avian paramyxovirus 1 isolate Asil/IR/AAA158/2019 fusion protein (F) gene | 100% | 0 | 99.37 | MN370894.1 |
| Avian paramyxovirus 1 isolate Ck/IR/EMMA140/2018 fusion protein (F) gene | 100% | 0 | 99.37 | MN370896.1 |
| Avian paramyxovirus 1 isolate Ck/IR/EMMA121/2018 fusion protein (F) gene | 100% | 0 | 99.37 | MN370895.1 |
| Avian paramyxovirus 1 strain Ck/IR/MAM72/2018 fusion protein gene | 100% | 0 | 99.16 | MH481362.1 |
| Avian paramyxovirus 1 strain Ck/IR/MAM68/2017 fusion protein gene | 100% | 0 | 99.16 | MH481361.1 |
| Avian paramyxovirus 1 strain Ck/IR/MAM31/2017 fusion protein gene | 100% | 0 | 99.16 | MH247185.1 |
| Avian orthoavulavirus 1 isolate Ph/IR/AMMM116/2018 fusion protein gene | 100% | 0 | 99.16 | MT627321.1 |
| Avian paramyxovirus 1 isolate ByCk/IR/MAME98/2017 fusion protein gene | 100% | 0 | 99.16 | MN786527.1 |
| Avian paramyxovirus 1 isolate Ck/IR/EMA92/2017 fusion protein gene | 100% | 0 | 99.16 | MN786517.1 |
| Avian paramyxovirus 1 isolate Ck/IR/EMA86/2017 fusion protein gene | 100% | 0 | 99.16 | MN786516.1 |
| Avian paramyxovirus 1 isolate Ck/IR/MAME85/2017 fusion protein gene | 100% | 0 | 99.16 | MN701086.1 |
| Avian paramyxovirus 1 strain Ck/IR/MAM18/2017 fusion protein gene | 100% | 0 | 99.16 | MN242824.1 |
| Avian paramyxovirus 1 strain Ck/IR/EMA148/2019 fusion protein (F) gene | 100% | 0 | 99.16 | MN481201.1 |
| Avian paramyxovirus 1 strain Ck/IR/EMA152/2019 fusion protein (F) gene | 100% | 0 | 99.16 | MN481192.1 |
| Avian paramyxovirus 1 strain Ck/IR/MAM55/2017 fusion protein gene | 100% | 0 | 98.95 | MH247187.1 |
Pathotyping of NDV isolates in chicken using MDT
To characterize the isolates biologically and clarify whether they are avirulent (vaccine strains) or virulent strains, the pathogenicity of NDV isolated from the chickens was assessed by Mean Death Times (MDT). The MDT observed in embryonated chicken eggs was 58hr for (Y03), 57hr for(Y11) and 59hr for (Y12). The post-inoculation chickens showed a diffusely red lesions in the subcutaneous tissue of the head and was filled with blood, edema in the head region and the blood vessels over the body were prominent, diffuse congestion and hemorrhage in the whole body.
To determine the ELD50, 10-fold serial dilutions of AF samples were used (i.e., 10-1, 10-2, 10-3 ,10-4, 10-5 and 10- 6). Each of these dilutions was inoculated into 5 embryonated chicken eggs. Mortality was recorded up to seven days post-inoculation. The calculated ELD50 of AF sample isolates was 10-8.839 to 10-8.990/0.1ml of virus (Table 3). Clinically, chicks in all groups except control group showed clinical signs ranged from general non-specific signs of depression, closed eyes, off intacking and lateral recumbence too nervous in nature including paresis and paralysis of legs (one or both) and wings (one or both), twisting of the neck, muscle tremors and incoordination.
The isolation and pathotyping of NDVs from the field of chickens is critical for the control of ND and vaccination evaluation because the commercial chickens were vaccinated, almost all flocks were accompanied by high mortality rates (25-90%) (Kapczynski and King, 2005). Clinical and PM findings of NDV isolates are recorded in the Table 1. In addition, the isolation of NDV from the organ samples collected from different localities was confirmed by HA and HI tests and real time PCR (Table 2). The virulence of NDV was measured by MDT, which varied from 57-59 hrs, showing virulence of field NDV according to OIE international standards. The NDV infection even in a well-vaccinated flock can occur because some of the birds will have a poor vaccine response and will be susceptible to infection. This attributed to ND vaccines that do not protect vaccinates from infection along with viral shedding. Also, parental immunity contributed to vaccination inefficiency in young chicks (Kapczynski and King, 2005).
Conclusions and Recommendations
In conclusion, our results indicated that the NDV isolates among vaccinated chickens were virulent and associated with endemic ND in poultry farms in Babylon province of Iraq in spite of extensive vaccination program. The NDV caused severe economic losses in most of these farms due to vaccination failure, inefficient vaccination or virus genetic drift should be considered to draft the efficiency of the commercially available vaccines against these isolates and multiple sources of vaccine. Finally, continuous checking of the flocks’ immunological status should be carried out after each vaccination to evaluate the antibody response to administrated vaccines with proper biosecurity practice.
Acknowledgement
Thank you very much to the Chairman of department of pathology and poultry disease at AL-Qasim green University.
Novelty Statement
Identification of newcastle disease viruses from naturally infected chickens is novel research work for using in future studies for embryonic health-related investigations.
Author’s Contribution
Yasser Asaad Hameed Al shareef and Firas Hussain Kadim Abawi are participated in designing, experimental works, drafting and final writing of manuscript with similar participation.
Ethics approval and consent to participate
The project was approved by the local ethical committee in Al-Qasim Green University, Babylon of Babylon, and received approval to conduct this scientific investigation in ethical-research comitee.
Conflict of interests
The authors have declared no conflict of interest.
REFERENCES
Aldous EW, Alexander DJ (2001). Detection and differentiation of Newcastle disease virus (avian paramyxovirus type 1). Avian Pathol., 30: 117-128. https://doi.org/10.1080/03079450120044515
Alexander DJ (1988). Newcastle disease Kluwer Academic Publishers, Boston. pp. 147-160. https://doi.org/10.1007/978-1-4613-1759-3_9
Alexander DJ (2000). Newcastle disease and other avian paramyxoviruses. Rev. Sci. Tech. Office Int. Des. Epizooties, 19(2): 443–455. https://anyflip.com/ffxh/wlnr/basic/ https://doi.org/10.20506/rst.19.2.1231
Amarasinghe GK, Aréchiga Ceballos NG, Banyard AC, Basler CF, Bavari S, Bennett AJ, Blasdell KR, Briese T, Bukreyev A, Caì Y, Calisher CH (2018). Taxonomy of the order mononegavirales: Update 2018. Arch. Virol., 163: 2283–2294. https://doi.org/10.1007/s00705-018-3814-x
Dimitrov KM, Abolnik C, Afonso CL, Albina E, Bahl J, Berg M, Briand FX, Brown IH, Choi KS, Chvala I, Diel DG (2019). Updated unified phylogenetic classification system and revised nomenclature for Newcastle disease virus. Infect. Genet. Evol., 74: 103917. https://doi.org/10.1016/j.meegid.2019.103917
Eid AAM, Hussein A, Hassanin O, Elbakrey RM, Daines R, Sadeyen JR, Abdien HMF, Chrzastek K, Iqbal M (2022). Newcastle disease genotype VII prevalence in poultry and wild birds in Egypt. Viruses, 14. https://doi.org/10.3390/v14102244
Ewies SS, Ali A, Tamam SM, Madbouly HM (2017). Molecular characterization of Newcastle disease virus (genotype VII) from broiler chickens in Egypt. Beni-Suef Univ. J. Basic Appl. Sci., 6(3): 232–237. https://doi.org/10.1016/j.bjbas.2017.04.004
Felsenstein J (1985). Confidence limits on phylogenies: An approach using the bootstrap. Evolution, 39: 783-791. https://doi.org/10.1111/j.1558-5646.1985.tb00420.x
Huang Z, Panda A, Elankumaran S, Rockemann DD, Samal SK (2004). The haemagglutination-neuranimidase protein of Newcastle disease virus determines tropism and Virulence. J. Virol., 78: 4176- 4184. https://doi.org/10.1128/JVI.78.8.4176-4184.2004
Jestin V, Jestin A (1991). Detection of Newcastle disease virus RNA in infected allantoic fluid by in vitro enzymatic amplification (PCR). Arch. Virol., 118: 151-161. https://doi.org/10.1007/BF01314026
Jukes TH, Cantor CR (1969). Evolution of protein molecules. In: (ed. H.M. Munro) Mammalian Protein Metabolism, Academic Press, New York. pp. 21-132. https://doi.org/10.1016/B978-1-4832-3211-9.50009-7
Kapczynski DR, King DJ (2005). Protection of chickens against overt clinical disease and determination of viral shedding following vaccination with commercially available Newcastle disease virus vaccines upon challenge with highly virulent virus from the California 2002 exotic Newcastle disease outbreak. Vaccine, 23(26): 3423-3433. https://doi.org/10.1016/j.vaccine.2005.01.140
Kumar S, Stecher G, Tamura K (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol., 33: 1870-1874. https://doi.org/10.1093/molbev/msw054
Liang R, Cao DJ, Li JQ, Chen J, Guo X, Zhuang FF, Duan MX (2002). Newcastle disease outbreaks in western China were caused by the genotypes VIIa and VIII. Vet. Microbiol., 87(3): 193-203. https://doi.org/10.1016/S0378-1135(02)00050-0
Mayo M (2002). A summary of taxonomic changes recently approved by ICTV. Arch. Virol., 147: 1655–1656. https://doi.org/10.1007/s007050200039
OIE World Organization for Animal Health (2008). Chapter 2.3.14 In: Manual of diagnostic tests and vaccines for terrestrial animals, 6th ed., OIE, Paris, France. pp. 576–589.
OIE (2012). Newcastle disease. In: OIE terrestrial manual: Manual of diagnostic tests and vaccines for terrestrial animals. World Organization for Animal Health. Paris. France.
Pazhanivel N, Balsubramaniam GA, George VT, Mohan B (2002). Study of natural outbreak of Newcastle disease in and around Namakkal. Indian J. Vet., 79(3): 293-294.
Piancenta AM, King DI, Seal BS, Zhang J, Brown CC (2006). Pathogenesis of Newcastle disease in commercial and specific pathogen free turkeys experimentally infected with isolates of different virulence. Vet. Pathol., 43: 168-178. https://doi.org/10.1354/vp.43-2-168
Saitou N, Nei M (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol., 4: 406-425.
Śmietanka KR, Minta ZE, Domańska-Blicharz KA (2006). Detection of Newcastle disease virus in infected chicken embryos and chicken tissues by RT PCR. Bull. Vet. Inst. Pulawy., 50: 3-7.
Spackman E, Senne DA, Bulaga LL, Myers TJ, Perdue ML, Garber LP, Lohman K, Daum LT, Suarez DL (2003). Development of real-time RT-PCR for the detection of avian influenza virus. Avian Dis., 47: 1079- 1082. https://doi.org/10.1637/0005-2086-47.s3.1079
Susta L, Miller PJ, Afonso CL, Brown CC (2011). Clinicopathological characterization in poultry of three strains of Newcastle disease virus isolated from recent outbreaks. Vet. Pathol., 48(2): 349–360. https://doi.org/10.1177/0300985810375806
Terregino C, Capua I (2009). Clinical traits and pathology of Newcastle disease infection and guidelines for farm visit and differential diagnosis BT - avian influenza and Newcastle disease: A field and laboratory manual. In: (eds. I. Capua and D.J. Alexander). Springer Milan, Milano, pp. 113–122. https://doi.org/10.1007/978-88-470-0826-7_9
Zhang L, Pan Z, Geng S, Chen X, Hu S, Liu H, Wu Y, Jiao X, Liu X (2010). Sensitive, semi-nested RT-PCR amplification of fusion gene sequences for the rapid detection and differentiation of Newcastle disease virus. Res. Vet. Sci., 89(2): 282–289. https://doi.org/10.1016/j.rvsc.2010.02.007
Zhao H, Wen Q, Wu Y, Zhang R, Liu X (2001). The relationship between HI’s antibody and virulent NDV infection in immunized chicken flocks. J. Yangzhan Univ., 4(2): 23-26. https://www.cabidigitallibrary.org/doi/full/10.5555/20013083343
To share on other social networks, click on any share button. What are these?






