Advances in Animal and Veterinary Sciences
Research Article
Optimization and Validation of a Diagnostic Real-Time PCR for Bovine Brucellosis
Falguni Mukherjee1a, Kommoju Nagmani2a, Kota Sri Naga Leela Surendra1, Bhaskaran Mohana Subramanian3, Vijay Sriram Bahekar1, Amitesh Prasad1, Samir Kumar Rana1, Ponnanna Nadikerianda Muthappa1, Girish Kumar Sharma4, Villuppananoor Alwar Srinivasan5*
1National Dairy Development Board, R&D Laboratory, Hyderabad, 500032, Telangana, India; 2Department of Biotechnology, Jawaharlal Nehru Technological University Hyderabad and Research and Development, Indian Immunologicals Limited, Hyderabad, 500032, Telangana, India; 3Translational Research Platform for Veterinary Biologicals, Tamil Nadu Veterinary and Animal Sciences University, Chennai, 600051, India; 4National Dairy Development Board, Anand, 388001, Gujarat, India; 5National Dairy Development Board, Animal Health, Gachibowli, Hyderabad 500032, Telangana, India.
Abstract | A diagnostic real-time PCR (qPCR) targeting the Brucella cell salt extractable outer membrane protein gene bcsp-31 was optimized for identification of genus Brucella. The assay had an analytical sensitivity of 30fg and reliably detected up to one copy number of the positive control plasmid construct, and 1x104 Brucella cells/reaction from spiked bovine tissue matrices. The qPCR detected DNA from 30 Brucella strains but not from non-Brucella strains. The qPCR was reliable, reproducible and could be completed in 72 minutes. Comparative quantification of Brucella copy number was established by utilizing normalized Cq values. The best return of validation estimates were obtained when animal-wise results of qPCR were compared to the combined status of culture and serology (n=230) since the two assays were strongly associated (κ =0.848 at 95% CI) and revealed a diagnostic sensitivity (DSe) of 77.8% and specificity (DSp) of 100%, positive and negative predictive (PPv and NPv) value of 100% and 94.61% at 95% CI, respectively. In contrasts, the DSe, DSp, PPv and NPv values obtained after comparison of results of qPCR and culture were 100%, 86.55%, 18.2% and 100% at 95% CI, respectively. Therefore, if the estimates were assessed in parallel, together they could form a reliable and rapid diagnostic tool for screening bovine brucellosis.
Keywords | Brucellosis, bcsp-31, qPCR, conventional PCR
Editor | Kuldeep Dhama, Indian Veterinary Research Institute, Uttar Pradesh, India.
Received | August 01, 2015; Revised | August 20, 2015; Accepted | August 20, 2015; Published | October 05, 2015
*Correspondence | Villuppananoor Alwar Srinivasan, National Dairy Development Board, Telecom Nagar, Gachibowli, Hyderabad, India; Email: [email protected]
aFalguni Mukherjee and Kommoju Nagmani contributed equally to this work.
Citation | Mukherjee F, Nagmani K, Surendra KSNL, Subramanian BM, Bahekar VS, Prasad A, Rana SK, Muthappa PN, Sharma GK, Srinivasan VA (2015). Optimization and validation of a diagnostic real-time PCR for bovine brucellosis. Adv. Anim. Vet. Sci. 3(11): 577-587.
DOI | http://dx.doi.org/10.14737/journal.aavs/2015/3.11.577.587
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2015 Mukherjee et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
Bacteria belonging to the genus Brucella cause bovine brucellosis characterized by abortion and infertility in cattle and buffaloes, resulting in economic loss to the dairy industry (OIE, 2014; Corbel, 1997). The disease exists world-wide except in some developed countries (Corbel, 1997). However, the disease is endemic in India (Renukaradhya et al., 2002; Mukherjee et al., 2005). Bovine brucellosis is conventionally diagnosed by serology; and isolation of Brucella species by culture is considered as the Gold standard (Corbel, 1997; Alton et al., 1975). Isolation by culture is time taking, biohazardous and requires Class-III containment facilities, whereas serological tests may not always indicate the true status of the disease and are sometimes affected by specificity and sensitivity issues (Young et al., 1991).
Detection of Brucella by molecular methods is an attractive alternative approach for diagnosis since it can identify the organism without culture (Yu and Nielsen, 2010). Earlier, conventional PCR was used for the detection of Brucella from culture and clinical samples targeting the bcsp31, 16S rRNA, 16S-23S intergenic transcribed spacers (ITS), IS711, per and omp2 genes (Baily et al., 1992; Romero et al., 1995; Rijpens et al., 1996; Henault et al., 2000; Bogdanovich et al., 2004; Leal-Klevezas et al., 1995). During the past 15 years quantitation of the Brucella genome from cultures and clinical samples has been reported using qPCR targeting the bcsp-31, IS711, 16S-23S spacer, omp25, per and omp31 genes employing SYBR Green labelled probes, hydrolysis probes or systems that use fluorescence resonance energy transfer for specific hybridization with DNA template (Redkar et al., 2001; Kattar et al., 2007; Queipo-Ortuno et al., 2008; Zhang et al., 2011). Most of these reports indicated above were based on clinical studies derived from samples from human cases, and the selected gene targets were bcsp 31 or the IS711 element. But fewer number of such studies have been conducted on animals or animal products (Amoroso et al., 2011; Sohrabi et al., 2011; Dehkordi et al., 2012; Sidor et al., 2013; El Behiry et al., 2014; Dean et al., 2014). However, reports on validation of qPCR for brucellosis are still scarce in the literature (Probert et al., 2004; Debeaumont et al., 2005; Amoroso et al., 2011; Sidor et al., 2013). Most of these reports are based on screening of DNA panels covering an array of Brucella and non-Brucella strains (Bogdanovich et al., 2004; Probert et al., 2004), but only a few on actual clinical samples from animal or human origin (Amoroso et al., 2011; Sidor et al., 2013). In recent times there are only two reports of a validated Brucella genus specific qPCR; one from marine mammals (Sidor et al., 2013) and another from human serum (Debeaumont et al., 2005). In present study we describe the validation of the qPCR assay for identification of genus Brucella from clinical samples of bovine and bubaline origin from India, as prescribed in the guidelines of the validation and quality control of polymerase chain reaction methods used for diagnosis of infectious diseases (OIE, 2008) and according to requirements of the minimum information necessary for evaluating qPCR experiments (Bustin et al., 2009).
MATERIALS AND METHODS
Source of Clinical Samples and Brucella Isolates
A total of 867 clinical samples comprising of blood (n=230), milk (n=141), nasal and vaginal swabs (n=222 each) were used in the study. The samples were collected from 262 cattle and 20 buffaloes from 6 different farms, from the states of Telangana and Gujarat in India. Semen samples (n=52) from cattle and buffaloes from the state of Gujarat, India were also included in the study. Twenty seven field isolates of Brucella from Telangana and Gujarat region were included along with three reference Brucella strain.
Source and Maintenance of the Reference and Field Strains of Bacteria and Virus
Details of the strains used in the study are furnished in the Table 1. Brucella reference and field strains were maintained as per standard protocol (Alton et al., 1988). Yersenia enterocolitica O:3 and O:9 were maintained on Brain Heart Infusion agar (BD, U.S.A) at 28oC, Vibrio cholerae Ogawa and Inaba strains were grown in Terrestrial Yeast Extract medium (BD, U.S.A) at 25oC. Mycobacterium avium subspecies paratuberculosis (MAP) was maintained in 7H9 broth (BD, U.S.A) at 37oC. Agrobacterium tumefaciens was maintained in Yeast Extract broth at 28oC overnight. E. coli was propagated on Luria Bertani agar (Himedia, India) at 37oC for 24 hours. Bovine herpesvirus-1 (BHV-1) culture propagated in MDBK cells (ATCC Cat. No. CCC-22TM) was obtained from the R&D facilities of Indian Immunologicals Limited, Hyderabad.
Extraction of DNA from Virus, Bacteria and Clinical Samples
DNA extraction from BHV-1, Brucella, other bacteria except MAP and clinical samples was done as per the ‘blood and body fluid protocol’ of Qiagen Blood mini kit, Germany with slight modifications which includes treatment with lysis buffer for 30 minutes at 56oC, extended treatment with ethanol for 20 minutes and final DNA elution in 50-75µl of TE buffer. In case of blood and milk samples post lysis spinning was done at 12000 rpm for 10 minutes so that the sample could easily pass through the spin column. MAP DNA extraction was done by Tetracore kit (Rockville, USA).
Selection of Gene Target, Primers and Taqman Probes
The bcsp-31 gene (Gene Bank accession number M20404) encoding 31kDa antigen of Brucella species was selected as the gene target. The primers and probe were designed using the software from Genscript (www.genscript.com/tools.html#biology). The sequence of the primers and probe is as follows:
bcsp31 forward primer: 5’CTCGGTTGCCAATATCAATG 3’;
bcsp31 reverse primer: 5’ATATGGATCGTTTCCGGGTA 3’;
bcsp31 probe: FAM 5’CCGGTGCCGTTATAGGCCCA 3’ TAMRA
The selected primers were expected to generate an amplicon of 165 bp in the qPCR.
Preparation of Plasmid Standards
The bcsp-31 gene was amplified by PCR from the DNA of Brucella vaccine strain RB51 using the primers B4 and B5 (Baily et al., 1992). The 223bp PCR product was purified and cloned into Topo vector pCRTM2.1-TOPO® (Topo cloning kit, Invitrogen, U.S.A) as per the manufacturer’s
Table 1: Specificity of the bcsp31 real time PCR
|
Name the of the species |
Source / Origin |
bcsp31 qPCR |
Cq Value (cut off 38) |
|
Brucella abortus 544 (23448) |
ATCC |
+ |
13.16 |
|
Brucella abortus S19 |
NDDB |
+ |
18.98 |
|
Brucella abortus RB51 |
Virginia Tech |
+ |
18.65 |
|
Yersenia enterolytica O:3 |
HAU |
– |
>40 |
|
Yersenia enterolytica O:9 |
HAU |
– |
>40 |
|
Vibrio cholerae ogawa |
NICE |
– |
>40 |
|
Vibrio cholerae Inaba |
NICE |
– |
>40 |
|
Bovine herpes virus isolate |
NDDB |
– |
>40 |
|
Mycobacterium avium paratuberculosis |
ATCC |
– |
>40 |
|
Agrobacterium tumifaciens |
ATCC |
– |
>40 |
|
E.coli DH5α |
ATCC |
– |
>40 |
|
Brucella Isolate 1 from milk |
NDDB |
+ |
16.85 |
|
Brucella Isolate 2 from milk |
NDDB |
+ |
16.33 |
|
Brucella Isolate 3 from milk |
NDDB |
+ |
16.1 |
|
Brucella Isolate 4 from milk |
NDDB |
+ |
16.48 |
|
Brucella Isolate 5 from milk |
NDDB |
+ |
16.00 |
|
Brucella Isolate 6 from milk |
NDDB |
+ |
16.3 |
|
Brucella Isolate 7 from milk |
NDDB |
+ |
16.21 |
|
Brucella Isolate 8 from milk |
NDDB |
+ |
17.95 |
|
Brucella Isolate 9 from milk |
NDDB |
+ |
15.58 |
|
Brucella Isolate 10 from milk |
NDDB |
+ |
15.58 |
|
Brucella Isolate 11 from milk |
NDDB |
+ |
27.20 |
|
Brucella Isolate 12 from milk |
NDDB |
+ |
15.55 |
|
Brucella Isolate 13 from milk |
NDDB |
+ |
18.81 |
|
Brucella Isolate 14 from milk |
NDDB |
+ |
20.20 |
|
Brucella Isolate 15 from milk |
NDDB |
+ |
21.18 |
|
Brucella Isolate 16 from milk |
NDDB |
+ |
20.81 |
|
Brucella Isolate 17 from milk |
NDDB |
+ |
17.35 |
|
Brucella Isolate 18 from milk |
NDDB |
+ |
16.13 |
|
Brucella Isolate 19 from milk |
NDDB |
+ |
17.81 |
|
Brucella Isolate 20 from milk |
NDDB |
+ |
18.98 |
|
Brucella Isolate 21 from milk |
NDDB |
+ |
19.02 |
|
Brucella Isolate 22 from milk |
NDDB |
+ |
19.23 |
|
Brucella Isolate 23 from milk |
NDDB |
+ |
19.10 |
|
Brucella Isolate 24 from milk |
NDDB |
+ |
18.23 |
|
Brucella Isolate 25 from milk |
NDDB |
+ |
18.15 |
|
Brucella Isolate 26 from milk |
NDDB |
+ |
20.20 |
|
Brucella Isolate 27 from milk |
NDDB |
+ |
19.92 |
ATCC: American Type Culture Collection, USA; HAU: Haryana Agricultural University, Hisar, India; NICE: National Institute of Cholera and Enteric Diseases, Kolkata, India; NDDB: National Dairy Development Board, Anand, India; USDA: United States Department of Agriculture
instructions. The resultant plasmid clone was used as standard construct for the qPCR. Based on the concentration and size of the plasmid construct (pCRTM2.1-TOPO® -Bru- bcsp31), copy number of the plasmid was determined. Formula for converting the DNA quantities into number of copies is as follows: (amount in ng x 6.022x1023) / (length in bp x 1x109ng/g x 650 g/ mole of bp). The plasmid standard was serially diluted to achieve a final plasmid copy number which ranged from 1x1010 to 1 copy per 5µl.
Optimization of Real Time Polymerase Chain Reaction
The assay was performed in Rotor Gene Q qPCR cycler (Qiagen, Germany) using Quantifast Taqman probe PCR master mix (Qiagen, Germany). The reaction was performed in 0.2ml PCR strip-tubes (Qiagen, Germany) with a total reaction volume of 25µl which contained 12.5µl of master mix and 10 picomoles of each primer, 10 picomoles of probe and 5µl of the template (containing serially diluted DNA ranging from 300 ng to 30 fg). Reaction conditions were set as follows: Hold at 95oC for 5 minutes, cycling at 95oC for 5 seconds and 60oC for 30 seconds consisting of 60 cycles. The positive standard construct was serially diluted from 1010 to 1 copy number and real time reaction was performed for each dilution of the standard. Cq values of the standards were plotted on a graph against the initial copy numbers of the plasmid and the reaction efficiency and correlation coefficient (R2 values) were determined. The sample quantification was performed by plotting the sample Cq values in the standard graph. Clinical samples (blood, milk, nasal/vaginal swabs and semen) from known Brucella culture negative and positive animals were used to determine the ideal cut-off threshold cycle values.
Analytical Specificity and Sensitivity
ASp of the assay was determined by using DNA from various reference bacteria and virus. ASe of the assay was determined by performing the assay on plasmid standards in triplicates with known initial copy numbers of 1x1010 to one copy number and also on DNA isolated from B.abortus 544 serially diluted in triplicates.
Repeatability and Reproducibility
The intra-assay repeatability was determined by performing the assay using positive plasmid controls serially diluted from 1x1010 to 1 copy number in triplicates. The inter-assay reproducibility of the assay was analyzed by testing positive plasmid controls on three different days.
Sample Matrix Studies
B.abortus 544 strain was serially diluted from 1x1010 to one colony forming unit (cfu) and each of these dilutions were spiked into various chemical and biological matrices - phosphate buffered saline, blood, milk, tryptic soya broth (BD,U.S.A) and semen in triplicates. DNA was extracted from all the samples and qPCR was performed.
Brucella Genome Quantification
Reference genes of host tissue are used as exogenous controls in qPCR for normalization, to nullify inter-assay variations. Here, an unrelated DNA which was spiked in equal quantity in the sample was used as exogenous control to normalize the data. Various negative sample matrices like skimmed milk, pasteurized milk, non-pasteurized milk, cattle blood, buffalo blood, nasal swab, vaginal swab, prepucial swab (in tryptic soya broth) and phosphate buffered saline were spiked with B.abortus 544 strain with 3.7x109 cfu in duplicates. The same samples were further spiked with 6x106 copies of unrelated, linearized plasmid DNA containing HPV18L1 gene of Human Papilloma Virus (HPV). The DNA was extracted and assayed for bcsp31 gene and HPV18L1 gene by qPCRs. The copy numbers of Brucella and HPV were calculated by two independent standard curves and the Brucella DNA copy numbers were normalized using the copy numbers of HPV DNA. The normalized copy number is the ratio of bcsp31: HPV18L1 copy numbers for a particular sample. The qPCR for HPV used in this study is an in house method. Sequence of primers and probe targeting the HPV18L1 gene (Gene bank accession number: AY383628.1) is as follows:
HPV forward: 5’-TGGAGACCATCCGATAACAC-3’;
HPV reverse: 5’- GGATGTCTTGTTTGTTTCCG-3’;
HPV probe: 5’-FAM/TCT GTG TTC ACC ACC CGG GC/TAMRA/-3’
Master mix, reaction volume and conditions were same as that of Brucella qPCR. HPV18L1 quantification was done using a serially diluted standard plasmid construct.
Brucella genome quantification was done in the similar way for 37 animals from two farms which were suspected for brucellosis.
Estimation of Diagnostic Sensitivity and Specificity
Analysis of the data was based on sample-wise and animal-wise treatment. Sets of 2 x 2 contingency tables were generated for comparing the results. In the first instance the data was compared to disease status by culture; and in the second instance compared to the combined status of culture and serology. An animal was considered positive for brucellosis if it was either positive by culture or serology. For serological analysis, a commercial ELISA kit (COMPELISA 400 RAI 2006, Animal Health Veterinary Laboratories Agency AHVLA, UK) was used for screening the animals (n=282). For isolation of Brucella species, 585 clinical samples (222 nasal and vaginal swabs each and 141 milk samples) originating from 282 animals were cultured employing modified Brucella Selective Media using 1X concentration of antibiotics cocktail as prescribed by Her et al. (2009).
DNA was extracted from 867 samples from the 282 animals (230 blood, 222 nasal swabs, 222 vaginal swabs, 141 milk and 52 semen samples) and were tested by qPCR and also compared by conventional PCR using the B4 and B5 primers (Baily et al., 1992). In the sample-wise approach all the above samples except for blood and semen were taken for culture isolation and the results were compared with qPCR for determining the DSe and DSp of the assay. In the animal-wise approach (n=230) the results were compared with (a) culture and with (b) the combined status of culture and serology.
Statistical Analysis
ASp of the assay on cultures of Brucella reference and field strains and organisms not belonging to genus Brucella from the laboratory repository was analyzed by a two tailed student t-test and by receiver operating characteristic (ROC) curve analysis (MedCalc® software version 14.12, 1993-2015). Intra-assay repeatability of the test was analyzed by determining the standard deviation (SD) between the three replicates of each sample. Inter-assay reproducibility of triplicate samples between runs on three different days were tested by measuring the SD using Bland Altman plot (MedCalc® software version 14.12, 1993-2015). DSe and DSp of qPCR with reference to culture results from clinical isolates was determined using ROC curve analysis (MedCalc® software ver 14.12, 1993-2015). The relative sensitivity of the qPCR and conventional PCR with reference to culture results was also analysed by ROC curve analysis (MedCalc® software version 14.12, 1993-2015) and kappa statistics (Graph Pad software).
RESULTS
Organisms serologically and phylogenetically related to Brucella like Yersenia enterolytica O:3, Yersenia enterolytica O:9, Vibrio cholrae Ogawa and Vibrio cholrae Inaba were negative in the qPCR assay. Also, the assay could not produce positive amplification from Bovine herpes virus-1, Mycobacterium avium subspeceis paratuberculosis, Agrobacterium tumefacians and E.coli DH5α. The assay detected B.abortus 544 strain, Brucella vaccine strain S19 and RB51. Twenty seven Brucella field isolates were having Cq values lesser or equal to 20 (Table 1). When a two tailed student t-test was done with the standards and negative controls, significant variation between the Cq values of negative and positive controls were detected (P<0.0001).
The assay could detect one copy number of the positive plasmid and 30 fg of B.abortus 544 DNA (Table 2). The mean Cq values obtained are depicted in the Table 2. To determine the cut off Cq value nuclease free water, phos
Table 2: Analytical sensitivity of the assay was detected by serially diluted positive plasmid and B.abortus 544 DNA
|
Positive standard-Plasmid construct |
||||||||
|
Copy number |
107 |
106 |
105 |
104 |
103 |
102 |
101 |
1 |
|
Triplicate Mean Cq Value |
8.5 |
12.2 |
19.2 |
22.8 |
26.3 |
29.7 |
33.1 |
35.6 |
|
SD |
0.83 |
1.23 |
0.42 |
0.48 |
0.38 |
0.43 |
0.56 |
0.87 |
|
SE |
0.27 |
0.41 |
0.14 |
0.15 |
0.12 |
0.14 |
0.18 |
0.28 |
|
95%CI |
8.0-9.0 |
11.4-13.0 |
18.9-19.5 |
22.5-23.1 |
26.1-26.5 |
29.4-30.0 |
32.7-33.5 |
35.1-36.1 |
|
Positive standard-Bacterial genomic DNA |
||||||||
|
DNA concentration |
300 ng |
30 ng |
3 ng |
300 pg |
30 pg |
3 pg |
300 fg |
30 fg |
|
Triplicate Mean Cq Value |
8.5 |
13.1 |
16.5 |
19.9 |
23.3 |
26.8 |
30.1 |
31.7 |
|
SD |
0.2 |
0.09 |
0.05 |
0.02 |
0.06 |
0.11 |
0.16 |
0.72 |
|
SE |
0.11 |
0.05 |
0.02 |
0.01 |
0.03 |
0.06 |
0.08 |
0.41 |
|
95%CI |
8.2-8.7 |
13.0-13.1 |
16.5-16.6 |
19.8-19.9 |
23.3-23.4 |
26.7-27.0 |
29.9-30.2 |
30.9-32.5 |
phate buffered saline (PBS), tryptic soya broth (BD, U.S.A and clinical samples such as blood, milk, nasal and vaginal swabs from animals with known Brucella negative status were included in the test; the Cq values for these samples were found to be between 38 to 45. Furthermore, the cut off cycle threshold for positive amplification was determined as 38 and the optimum number of amplification cycles for the assay were fixed as 40. The signal obtained for any test sample around 38 and above were considered non-specific. Standard graph plotted using the ten-fold serial diluted plasmid standards displayed linearity up to 1 copy as the lowest limit of quantification. Repeated runs with these standards exhibited a significant co-efficient of correlation (R2 value) ranging between 0.94 to 0.99 and reaction efficiency ranging from 97 to 99 %. Hence, this standard curve was used to ascertain the number of copies of target DNA present in the samples under test.
The intra-assay analysis for repeatability (Figure 1) and inter-assay analysis for reproducibility (Figure 2) showed that the values for standard deviation (SD) lay within the acceptable range of mean ± 1.96 SD.
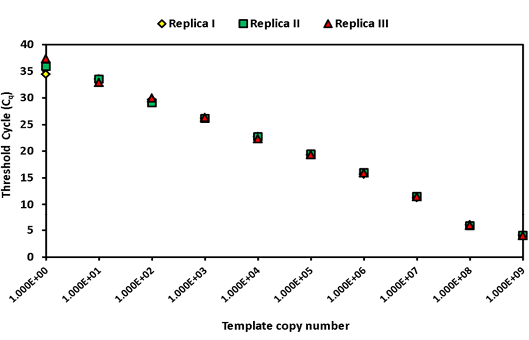
Figure 1: Intra assay variability wherein three replicates of serially diluted positive plasmid runs on the same day
The studies indicated that up to 1x104 cfu of spiked B. abortus 544 cells could be reliably detected in various samples matrices; the Cq values showed a linear order from 1x 107 to 1x104 cfu (Table 3).
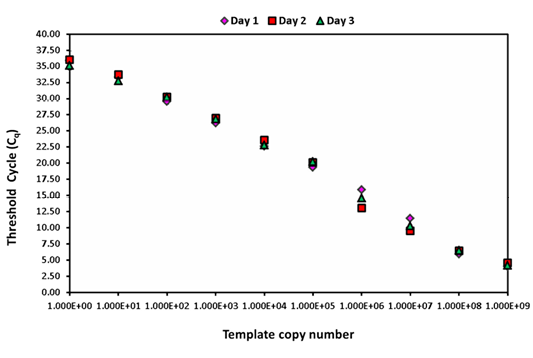
Figure 2: Inter assay variability wherein serially diluted positive plasmid runs on three different days
Table 3: Mean Cq values of clinical samples spiked and serially diluted with B.abortus 544 strain
|
B. abortus 544 strain cfu/ml spiked |
Mean of triplicates of Cq values of different clinical matrices spiked |
||||
|
PBS |
Blood |
Milk |
TSB |
Semen |
|
|
107 |
25.8 |
25.1 |
25.4 |
25.4 |
25.3 |
|
106 |
29.6 |
29.1 |
29.3 |
29.3 |
29.2 |
|
105 |
32.8 |
31.2 |
32.0 |
32.0 |
31.7 |
|
104 |
34.1 |
35.6 |
34.9 |
34.9 |
35.1 |
|
103 |
35.0 |
36.7 |
35.9 |
35.9 |
36.1 |
|
102 |
35.5 |
36.2 |
35.9 |
35.9 |
36.0 |
|
101 |
34.3 |
35.4 |
34.9 |
34.9 |
35.0 |
|
1 |
36.6 |
35.3 |
24.3 |
32.0 |
30.5 |
Table 4: Copy number of bcsp31 genome detected in clinical samples by qPCR
|
S.No |
Animal ID |
Sample type |
Serology |
Culture isolation |
qPCR result |
||
|
Cq Value |
Result |
Normalized Brucella copy number |
|||||
|
1 |
50 |
Nasal swab |
Negative |
Negative |
N |
Negative |
0.00E+00 |
|
2 |
2 |
Nasal swab |
Negative |
Negative |
35.7 |
Positive |
2.16E-04 |
|
3 |
11 |
Nasal swab |
Negative |
Negative |
N |
Negative |
0.00E+00 |
|
4 |
8 |
Nasal swab |
Negative |
Negative |
35.2 |
Positive |
2.14E-03 |
|
5 |
17 |
Nasal swab |
Positive |
Positive |
35.4 |
Positive |
2.15E-03 |
|
6 |
10 |
Nasal swab |
Negative |
Negative |
N |
Negative |
0.00E+00 |
|
7 |
13 |
Nasal swab |
Negative |
Positive |
36.3 |
Positive |
8.50E-04 |
|
8 |
367484 |
Nasal swab |
Positive |
Negative |
37.3 |
Positive |
2.65E-03 |
|
9 |
367472 |
Nasal swab |
Negative |
Negative |
37.1 |
Positive |
1.05E-03 |
|
10 |
367513 |
Nasal swab |
Positive |
Negative |
36.9 |
Positive |
1.04E-03 |
|
11 |
367499 |
Nasal swab |
Negative |
positive |
35.2 |
Positive |
2.13E-03 |
|
12 |
367492 |
Nasal swab |
Positive |
Negative |
N |
Negative |
0.00E+00 |
|
13 |
367511 |
Nasal swab |
Negative |
Negative |
37 |
Positive |
1.49E-03 |
|
14 |
367497 |
Nasal swab |
Negative |
Negative |
N |
Negative |
0.00E+00 |
|
15 |
367505 |
Nasal swab |
Negative |
Negative |
N |
Negative |
0.00E+00 |
|
16 |
367471 |
Nasal swab |
Negative |
Negative |
N |
Negative |
0.00E+00 |
|
17 |
367475 |
Nasal swab |
Negative |
Negative |
N |
Negative |
0.00E+00 |
|
18 |
367482 |
Nasal swab |
Negative |
Negative |
N |
Negative |
0.00E+00 |
|
19 |
367507 |
Nasal swab |
Negative |
Negative |
N |
Negative |
0.00E+00 |
|
20 |
367493 |
Nasal swab |
Negative |
Negative |
N |
Negative |
0.00E+00 |
|
21 |
367516 |
Nasal swab |
Negative |
Negative |
N |
Negative |
0.00E+00 |
|
22 |
367518 |
Nasal swab |
Negative |
Negative |
N |
Negative |
0.00E+00 |
|
23 |
367490 |
Nasal swab |
Positive |
Negative |
N |
Negative |
0.00E+00 |
|
24 |
367503 |
Nasal swab |
Positive |
Negative |
N |
Negative |
0.00E+00 |
|
25 |
367476 |
Nasal swab |
Positive |
Negative |
37.3 |
Positive |
2.17E-04 |
|
26 |
367478 |
Nasal swab |
Positive |
Negative |
N |
Negative |
0.00E+00 |
|
27 |
367500 |
Nasal swab |
Positive |
Negative |
N |
Negative |
0.00E+00 |
|
28 |
367473 |
Nasal swab |
Positive |
Negative |
N |
Negative |
0.00E+00 |
|
29 |
367470 |
Nasal swab |
Positive |
Negative |
36 |
Positive |
5.88E-04 |
|
30 |
367506 |
Nasal swab |
Negative |
Negative |
N |
Negative |
0.00E+00 |
|
31 |
16 |
Milk |
Positive |
Positive |
37.5 |
Positive |
2.54E-05 |
|
32 |
17 |
Milk |
Positive |
Positive |
36.2 |
Positive |
1.22E+03 |
|
33 |
13M |
Milk |
Positive |
Positive |
38 |
Positive |
5.51E-06 |
|
34 |
1 |
Milk |
Positive |
Positive |
32.3 |
Positive |
1.00E-02 |
|
35 |
3 |
Milk |
Positive |
Positive |
37 |
Positive |
1.60E-02 |
|
36 |
1 |
Vaginal swab |
Positive |
Positive |
37.5 |
Positive |
7.59E-03 |
|
37 |
3 |
Vaginal swab |
Positive |
Positive |
34.1 |
Positive |
4.71E-02 |
N=Negative
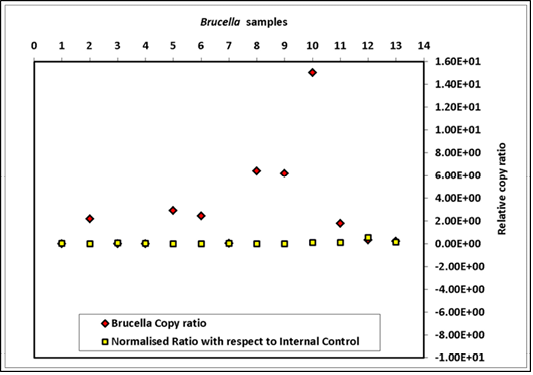
The genome quantification, derived from spiking a total of nine sample matrices equally with 3.7x109 cfu of B. abortus 544 and with 6x106 copies of exogenous control DNA of HPV18L1 are summarized in Figure 3. The results indicated that normalization with extraneous DNA was essential, as variation in Cq value was 103 folds higher without normalization (Figure 3). The R2 values for bcsp31 qPCR ranged from 0.94 to 0.99 and for the HPV 0.97 to 0.99. The PCR efficiencies for both the assays varied between 97-99%. Further, the effect of normalization on Cq value of qPCR
Table 5: qPCR, culture Isolation and conventional PCR results for clinical samples
|
Farm ID |
Blood |
Vaginal swab |
Nasal swab |
Milk |
|||||||||||
|
Total |
qPCR |
PCR |
Total |
qPCR |
Isolation |
PCR |
Total |
qPCR |
Isolation |
PCR |
Total |
qPCR |
Isolation |
PCR |
|
|
Farm 1 |
17 |
0 |
0 |
17 |
2 |
0 |
0 |
17 |
1 |
0 |
0 |
17 |
3 |
0 |
3 |
|
Farm 2 |
15 |
0 |
0 |
15 |
5 |
0 |
5 |
15 |
3 |
0 |
0 |
15 |
1 |
0 |
0 |
|
Farm 3 |
51 |
2 |
0 |
51 |
25 |
4 |
6 |
51 |
26 |
4 |
10 |
34 |
24 |
4 |
4 |
|
Farm 4 |
29 |
2 |
2 |
21 |
7 |
0 |
1 |
21 |
12 |
1 |
8 |
8 |
4 |
1 |
1 |
|
Farm 5 |
118 |
1 |
0 |
118 |
35 |
0 |
0 |
118 |
18 |
1 |
0 |
67 |
22 |
0 |
0 |
|
Grand Total |
230 |
5 |
2 |
222 |
74 |
4 |
12 |
222 |
60 |
6 |
18 |
141 |
54 |
5 |
8 |
|
% Positivity |
2.17 |
0.86 |
33.33 |
1.8 |
5.4 |
27.02 |
2.7 |
8.1 |
38.29 |
3.54 |
5.67 |
||||
Table 6: Animal wise result of serology, qPCR and culture isolation
|
Farm ID |
Total animals |
Serology |
qPCR (Positive in any one of the sample of each animal - NSa/VSb/Milk/Blood) |
Cultural Isolation (positive in any one of the sample of each animal - NSa/VSb/Milk) |
|
Farm 1 |
17 |
1 |
1 |
0 |
|
Farm 2 |
15 |
7 |
5 |
0 |
|
Farm 3 |
51 |
14 |
10 |
5 |
|
Farm 4 |
29 |
15 |
12 |
1 |
|
Farm 5 |
118 |
11 |
9 |
1 |
|
Farm 6 |
52 |
0 |
0 |
0 |
|
Total |
282 |
48 |
37 |
7 |
aNasal swab; bVaginal swab
Brucella copy ratio = copy number of sample before normalization/copy number of sample with lowest copy before normalization; Normalised ratio = copy number of sample after normalization/copy number of sample with lowest copy after normalization
for 37 clinical samples from two different farms compared to results of isolation by culture is furnished in Table 4.
The Cq values for DNA templates originating from milk samples varied from 37 to 38, for nasal swabs 35 to 39, for vaginal swabs 38, for blood 32.4 to 38, and for semen more than 38. The ROC analysis of the results of qPCR on 585 clinical samples compared to culture indicated that the assay had a DSe of 100% (95% CI = 78.2 - 100) and DSp of 70.0% (95% CI = 65.7 – 73.4). The comparative diagnostic estimates for individual clinical matrix are provided in Table 5. When animal-wise results (n=230) of qPCR (Table 6) were compared to culture alone the assay showed a fair degree of agreement (κ = 0.281; SE of κ = 0.085; 95% CI = 0.015 – 0.448). The comparative assessment of assays returned a DSp of 86.55% (95% CI = 81.35 – 90.73) and a DSe of 100% (95% CI = 58.93 – 100.0); a Positive Predictive value (PPv) of 18.92% 95% CI = 8.00 – 35.16) and Negative Predictive value (NPv) of 100% (95% CI = 98.08 – 100.0) (Table 7). However, when the same were compared with the combined status of culture and serology, the assays showed a very strong degree of agreement (κ = 0.848; SE of κ = 0.044 at 95% CI = 0.761 – 0.935). Moreover, the DSp was 100% (95% CI = 98.09 – 100.0) and DSn 77.08% (95% CI = 62.88 – 87.95. The PPv and NPv of the assay were 100% (95% CI= 90.42 – 100.0) and 94.61% respectively (95% CI = 90.55 - 92.77) (Table 7).
Relative sensitivity qPCR when compared to conventional PCR was found to be 100% (95% CI = 91.19 - 100) and the Sp was 80.26% (95% CI=77.28 - 83.01%) and the two tests showed a fair degree of agreement (κ =0.285). Blood and semen samples were not tested by culture in this ex
Table 7: Animal wise studies - correlation for qPCR, culture and serology
|
Correlation for qPCR Vs culture (Refer Table 6) |
Culture |
Total |
Kappa Value |
||
|
Positive |
Negative |
||||
|
qPCR |
Positive |
7 |
30 |
37 |
0.281 |
|
Negative |
0 |
193 |
193 |
||
|
Total |
7 |
223 |
230 |
||
|
Correlation for qPCR Vs combined culture and serology (Refer Table 6) |
Culture and Serology |
Total |
Kappa Value |
||
|
Positive |
Negative |
||||
|
qPCR |
Positive |
37 |
0 |
37 |
0.848 |
|
Negative |
11 |
193 |
204 |
||
|
Total |
48 |
193 |
241 |
||
periment. All samples positive by conventional PCR were positive by qPCR. All samples negative by qPCR were also negative by conventional PCR (Table 5).
DISCUSSION
The bcsp31 gene selected for this study is highly conserved among the species of the genus Brucella and most frequently used gene target for diagnosis of human brucellosis (Al Dahouk et al., 2007; Navarro et al., 2002; Morata et al., 2003); and therefore could potentially detect B. abortus, B. melitesnsis and B. suis strains that has been reported so far from cattle and buffaloes (Bricker et al., 1988; OIE, 2014). In our earlier findings (Mukherjee et al., 2007), compared to omp2 and 16S rRNA, the bcsp31 PCR was found to be 100% specific and was the most sensitive assay with PPv of 100% and NPv of 88%. Also numerous reports mentioned the use of bcsp31 for specific identification of genus Brucella from seropositive, active, relapsing, chronic cases in humans (Kattar et al., 2007; Mitka et al., 2007; Queipo-Ortuno et al., 2008). Recently the same gene target has been used specifically to detect Brucella in human serum, blood and cerebro-spinal fluid (Debeaumont et al., 2005; Colmenero et al., 2011; Sohrabi et al., 2011), in buffalo milk (Amoroso et al., 2011) and in clinical tissues from seals (Sidor et al., 2013).
The qPCR was specific since it did not amplify DNA from any non-Brucella templates. The limit of detection (LOD) for B.abortus 544 DNA was 30fg in the present assay and was comparable to earlier reports (Sidor et al., 2013; Probert et al., 2004). The estimated cut off Cq value was 38. The Cq<38 was declared as positive cut-off values for qPCR for human and camel serum samples (Sohrabi et al., 2011) and Cq<40 for testing an assay on a panel consisting of Brucella and non-Brucella DNA (Al Dahouk et al., 2007). The linear range for internal amplification control (positive plasmid construct), bacterial DNA, and bacteria spiked in various clinical matrix (bovine blood, milk, semen) were over 7, 7 and 3 orders of magnitude, respectively (2x102 – 2x109, 2x104 – 2x1011, and 2x102 – 2x 105 copies /ml, respectively). This range was comparable to the earlier reports (Debeaumont et al., 2005; Colmenero et al., 2011). The estimated SD for repeatability and reproducibility were within the acceptable range (OIE, 2014). The present assay had a PCR reaction efficiency varying from 97 to 99% which is similar to another report published by Debeaumont et al. (2005), using the bcsp31 on the B.melitensis template. The employment of the exogenous single copy gene HPV18L1 for co-spiking with sample DNA resulted in the normalization of Cq values as evidenced by reduction of variation in Brucella copy number by 100 folds.
Many reports have been published regarding the diagnostic estimates (DSe, DSp) of qPCR assays using the bcsp31 genome on human samples (Colmenero et al., 2011; Sohrabi et al., 2011; Sanjuan-Jimenez et al., 2013). The bcsp31 has been exploited for screening serum samples in camel (El Behiry et al., 2014), the IS711 (Gwida et al., 2011), the BMEII_0466 for identification of B. meltensis and BruAb2_0168 for B.abortus from aborted materials of cattle, buffaloes, camel, caprines and ovines (Dehkordi et al., 2012). In all the above reports the DSe appears to vary from 72% to 100% and the DSp, except in one report (Gwida et al., 2011) was 100%. However, none of these assays provide the complete estimates of validation. The validation reports of qPCR assays furnished by Debeaumont et al. (2005), Surucuoglu et al. (2009), on human samples, and those presented by Amoroso et al. (2011) on buffalo milk samples, and Sidor et al. 2013 on clinical samples from seals are therefore rare. Their studies had indicated that the DSe could vary from 64.7% to 88% and DSp 98.3% to 100%. Previously the estimates of diagnostic qPCR for brucellosis had been calculated based on case-wise (Surucuoglu et al., 2009) and also on sample-wise status in humans and in animals (Debeaumont et al., 2005; Sanjuan-Jimenez et al., 2013; Sohrabi et al., 2014), and on comparison with status by culture (Amoroso et al., 2011; El Behiry et al., 2014), serology (Gwida et al., 2011; Menshawy et al., 2014; Sohrabi et al., 2014) and combined status by culture and serology (Menshawy et al., 2014; Sohrabi et al., 2014).
In the present study, we have optimized and validated the diagnostic estimates of qPCR by animal-wise and sample-wise approach comparing the culture status in the first instance and the combined culture and serology status in the second instance. The multiple approach adopted for derivation of diagnostic estimates led to interesting observations. Using animal-wise approach and culture as reference the qPCR had a DSe of 100%, DSp 70%, PPv of 55.6% and NPv of 100%. In terms of DSe it was superior to the two validated qPCRs for detection of genus Brucella (DSe 64.71% - Debeaumont et al. (2005); DSe – 70.4% Sidor et al. (2013)) but in terms of DSp the estimate was inferior (DSp 70%) compared to other reports (DSp 100% and 98.3% as reported by Debeaumont et al. (2005) and Sidor et al. (2013), respectively). We calculated the PPv (55.6%) and NPv (100%) estimates of our assay that were not mentioned in the two validation reports cited above. The animal-wise and combined culture and serology approach significantly altered the estimates improving the DSp (76.4%) and PPv (77.78%) but lowering the DSe (70.47) and NPv (68.4%). A sample-wise estimation with respect to culture had returned similar diagnostic estimate where the DSe was 100% and DSp 70%. Nasal swabs (n=222) seemed to be the best sample template, because it returned a DSe of 100% and a DSp of 75%; also the nasal swab is easy to sample and is a non-invasive method. The animal-wise and culture status approach provided a further improvement in all the estimates (DSe 100%, DSp 86.55%, NPv 100%) of the assay except for a significant reduction in the PPv (18.92%). The best return of diagnostic estimate was derived from the animal-wise and combined culture and serology approach adopted for comparison, wherein except for lowering of the DSe (77.08%), the DSp, PPv and NPv were > 95% (DSp 100%, PPv 100%, PNv 94.61%). Also the two evaluation tools (qPCR vs culture and serology combined) reflecting the true status of the disease were very strongly associated (κ = 0.848). These diagnostic estimates derived from the present studies were therefore better from the two earlier reports (refer to the estimates cited above) on clinical samples from human (Debeaumont et al., 2005) and seals (Sidor et al., 2013).
Earlier reports had indicated that the relative sensitivities and specificities of qPCR, culture and serological assays may vary under various clinical settings. Thus some proportion of samples that were positive by culture was negative by qPCR (Debeaumont et al., 2005), similarly samples positive by IS711 PCR assay have been shown negative by culture (Sanjuan-Jimenez et al., 2013). Choice of media selected for isolation by culture may also affect sensitivity (Her et al., 2009; Sohrabi et al., 2014; Dean et al., 2014). The range of Cq values of clinical samples positive for brucellosis reported in the current study were similar (Cq - 33.3±4.6) to those reported earlier (Colmenero et al., 2011). The average concentration of DNA templates from clinical samples used for the assay was 50ng. Thus the presence of low copy number of bcsp31 in most of the clinical samples close to LOD was detectable in our assay. Presence of Brucella in low copy numbers in clinical samples from humans and seals has been reported earlier (Colmenero et al., 2011; Sidor et al., 2013).
Further, we had used B4 and B5 primers for conventional PCR in this study for comparing with qPCR that used primers in the assay that were different from B4 and B5, still when the assays were compared all samples positive by conventional PCR were also positive by qPCR. Also all samples negative by qPCR were negative by conventional PCR as well.
The diagnostic estimates of qPCR derived after normalization of Cq values of clinical templates were of a limited sample size. The accuracy of estimates could be improved if experiments were conducted on a larger sample size. However, the diagnostic estimates of the qPCR presented in this study can be applied in parallel for accurate diagnosis for ‘ruling in’ or ‘ruling out’ brucellosis in a bovine population. Since for ‘ruling out’ of the disease a test with at least 95% sensitivity and 75% specificity is required; and for confirmation of the disease, a test with at least 95% specificity and 75% sensitivity is required (Fegan et al., 1999). The present study has the potential to be used as a diagnostic tool or for conducting pre-vaccine survey of brucellosis status. This may even be evaluated to assess the therapeutic efficacy of Brucella vaccines by periodically estimating reduction in copy numbers following vaccination of infected animals.
ACKNOWLEDGEMENT
The authors are grateful to the management of the National Diary Development Board (NDDB), Anand, Gujarat for providing the facilities to carry out this work. Nagmani K expresses her gratitude to the Indian Immunologicals Limited, Hyderabad, India for providing the opportunity to work on the above topic with respect to partial fulfillment for her PhD thesis.
Authors Contribution
FM conceptualized and designed the study, conducted statistical analysis, and wrote the paper. KN carried out all the optimization and validation work, prepared the data and wrote the initial draft of the paper. KSNLS did the statistical analysis, plotted graphs, and edited the initial draft of the manuscript. BMS helped in designing of primers and TaqMan probes for Real Time PCR assays and was involved in the optimization of the qPCR assays, he also edited the initial draft of the paper. VSB and AP collected the clinical samples from the field. Field samples from Gujarat were collected by SKR. NMP provided the clone containing the positive plasmid construct of HPV-E6 and primer and TaqMan probes for qPCR. GKS provided the administrative and financial support from the NDDB. All the authors are thankful to VAS for critically reviewing and editing the manuscript.
Conflict of Interest
The authors declare no conflict of interest related to this article.
REFERENCES