Advances in Animal and Veterinary Sciences
Research Article
Advances in Animal and Veterinary Sciences 2 (1S): 26 – 31Special Issue–1 (Infectious Diseases of Animals and Global Health)
Therapeutic Potential of a Candidate relA Deletion Mutant Vaccine of Mycobacterium Avium Subsp. Paratuberculosis
Kun Taek Park, William C. Davis*
- Department of Veterinary Microbiology and Pathology, College of Veterinary Medicine, Washington State University, Pullman, WA 99164
*Corresponding author: [email protected]
ARTICLE CITATION:
Park KT, Davis WC (2014). Therapeutic potential of a candidate relA deletion mutant vaccine of Mycobacterium avium subsp. paratuberculosis. Adv. Anim. Vet. Sci. 2 (1S): 26 – 31.
Received: 2014–02–14, Revised: 2014–03–13, Accepted: 2014–03–15
The electronic version of this article is the complete one and can be found online at
(
http://dx.doi.org/10.14737/journal.aavs/2014/2.1s.26.31
)
which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited
ABSTRACT
Analysis of the immune response to a mutant of Mycobacterium avium subsp. paratuberculosis with a deletion of relA (MapΔrelA) revealed that deletion of relA interrupts the pathways used by Map to subvert the immune response in goats and cattle to establish a persistent infection. The immune response elicited by MapΔrelA cleared infection and limited the capacity of wild type Map to establish an infection. The findings indicate the mutant should be useful as a vaccine under field conditions. The present study was conducted to explore the therapeutic potential of MapΔrelA using a cow with late stage paratuberculosis. The cow was tested negative using Map skin test and peripheral blood mononuclear cells were unresponsive to antigen stimulation ex vivo. A strong CD4 and CD8 proliferative response was detected one week post vaccination with MapΔrelA. Analysis of the cytokine profile for four weeks post vaccination revealed a mixed response with up–regulation of pro–inflammatory and inhibitory cytokine gene expression. The findings indicate further testing of the mutant as a candidate vaccine should include examining analysis of the cellular events associated with restoring a protective immune response as well as its therapeutic potential.
INTRODUCTION
Various approaches have been taken to develop a vaccine to Mycobacterium avium subsp. paratuberculosis (Map) that induces sterile immunity against Johne’s disease (JD) or prevents disease progression. These approaches have included development of killed whole vaccines, live attenuated, DNA and subunit vaccines and tests in different animal species (Bull et al. 2007; Park et al. 2008; Juste et al. 2009; Kathaperumal et al. 2009; Scandurra et al. 2010; Bastida and Juste 2011; Stabel et al. 2012; Faisal et al. 2013). Although none of the vaccines induced sterile immunity they did slow disease progression and demonstrated that development of a vaccine that induces sterile immunity is feasible (reviewed in (Rosseels and Huygen 2008). Of interest, the studies also revealed the current vaccines can be used therapeutically to reverse immunopathology (Bull et al. 2007; Juste et al. 2009; Singh et al. 2010), an observation first observed by Koch (reviewed in (Burke 1993)). If animals are not too far advanced in clinical disease, the effect can be dramatic with a reversal of pathology (Singh et al. 2010). Unfortunately, the effect is transient as discovered by Koch (Burke 1993) and noted by Singh el al. (personal communication). The heightened immune response wanes allowing clinical disease to recur. Further studies are needed to determine what aspect of the immune response to Map accounts for the therapeutic efficacy and identify a method that increases the longevity of the therapeutic effect. One approach is to use an attenuated vaccine that minimizes some of the disadvantages associated with the use of killed vaccines. We have developed a deletion mutant that provides an opportunity to explore the therapeutic potential of an attenuated candidate live vaccine. Studies in calves and goats showed deletion of relA, a global regulator, interrupts the pathways used by Map to establish a persistent infection without reducing its immunogenicity. The mutant, MapΔrelA, elicits an immune response that clears infection and limits colonization of bacteria in challenge experiments (Park et al. 2011). The immune response is clearly different than the response elicited by wild type Map. The basis for this difference remains to be elucidated. The findings in the present study show the mutant offers an opportunity to characterize the components of the immune response associated with the therapeutic effect of vaccinating animals at the clinical stage of infection.
MATERIALS AND METHODS
Animals
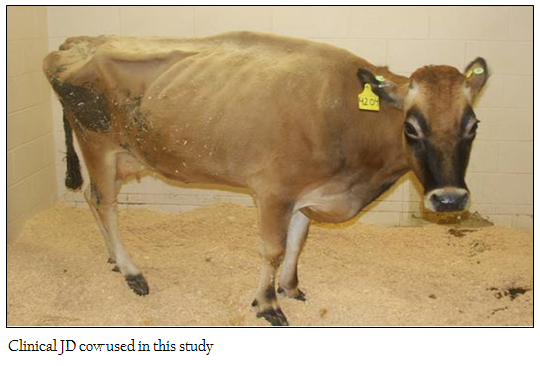
A cow infected with Map that was scheduled for disposal was obtained from a local dairy. The cow had lost weight and had continuous bouts of diarrhea and shedding bacteria. Clinical evaluation revealed some edema in the jaw and brisket (Figure 1). Analysis of the immune status showed that the cow was ELISA positive, skin test negative and unresponsive to antigen stimulation ex vivo as detected by flow cytometric (FC) analysis. The cow collapsed at 6 weeks after starting the experiment and was euthanized. Necropsy revealed the intestine had deteriorated.
Experimental Design
The cow was vaccinated with 109 of MapΔrelA (in 0.5 ml PBS) on the left side of neck behind ear by intra dermal injection. Blood was collected through the jugular vein at day 0 and 1, 2, 3, and 4 wk post vaccination (PV). Feces were collected on the same schedule as blood collection. The blood and feces were used as described below.
All animal treatments and experimental procedures were approved by the Washington State University Institutional Animal Care and Use Committee, USA.
Detection of Map DNA from Blood Samples by Real Time PCR
Ten milliliter of blood was mixed with 40 ml of red blood cell (RBC) lysis buffer (0.87 % Ammonium Chloride, 0.012 % Tris–Base, pH 7.3) in a 50 ml tube, and incubated at 37° C for 15 min. After RBC lysis, the leukocytes were pelleted by centrifugation at 600 xg for 10 min. The pellet was washed with 50 ml of phosphate buffered saline (PBS, pH 7.3), re–suspended in 1 ml of PBS, and transferred in to a 2 ml screw cap tube. Following centrifugation DNA was extracted with DNeasy Blood & Tissue kit with mechanical disruption as previously described (Park et al. 2011). The DNA extraction was done in duplicate at each bleeding time. A real time PCR (RT–PCR) method targeting IS900 sequence described by Irenge et al. (Irenge et al. 2009) was used to detect Map DNA from blood samples. Each DNA sample was run in duplicate.
Quantification of Bacterial Shedding in Feces by RT–PCR
The feces collected at each time point were processed for DNA extraction as previously described (Park et al. 2011). RT–PCR targeting Map–specific single copy gene, F57, was used for a correct quantification (Schonenbrucher et al. 2008). DNA extraction was done in duplicate, and the RT–PCR for each DNA sample was conducted in duplicate. A standard curve was generated using Map gDNA with 10 fold serial dilutions, and used for calculating the number of Map in the samples.
Peripheral Blood Mononuclear Cell Isolation and Stimulation with Map
Peripheral blood mononuclear cells (PBMC) from blood were isolated and stimulated with live Map in 6 well plates for 6 days as previously described (Allen et al. 2009; Park et al. 2011). Total RNA from PBMC was collected on day 3 for analysis of cytokine expression. Live PBMC were collected on day 6 for FC analysis.
Flow Cytometric Analysis
The same gating strategy, as used in previous studies, was used to analyze the CD4 and CD8 T cell proliferative response to live Map stimulation (Allen et al. 2009; Park et al. 2011). In brief, fresh PBMC (time 0) and stimulated PBMC with/without live Map (day 6) were labeled with three color combinations of monoclonal antibodies, CD4 (ILA11A, IgG2a), CD45R0 (ILA116A, IgG3), and CD25 (CACT116A, IgG1), or CD8 (7C2B, IgG2a), CD45R0, CD25 (Washington State University Monoclonal Antibody Center, Pullman, WA, USA). As shown in Figure 2, three electronic gates were used to analyze the data. The first two gates were used in side (SSC) and forward (FSC) light scatter to identify and color code resting (G1, non–proliferating, orange) cells and activated (G2, proliferating, blue) cells (Figures. 2A, 2D, 2G). Cells in G2, at T0, included monocytes and large lymphocytes. At day 6, G2 contained activated proliferating T cells. The third gate was placed on CD4 or CD8 positive cells to electronically isolate the cells for analysis. The results are displayed in FSC vs CD45R0 to show the relative frequency of naïve cells (resting cells, orange), and activated (blue) memory CD4 (Figure 2B, 2E, 2H) and CD8 (Figure 2C, 2F, 2I) cells. A FACSort flow cytometer and Cell Quest software (Becton Dickinson Immunocytometry Systems, San Jose, CA) were used to collect data. FCS Express software ver. 3 (De Novo software, Los Angeles, CA) was used to analyze the data.
Analysis of Cytokine Expression Profile
Quantitative reverse–transcription RT–PCR (qRT–PCR) was used to compare the relative cytokine transcription of PBMC stimulated with live Map at different time points. Total RNA was extracted from PBMC stimulated with live Map using TRIzol (Life Technologies, CA). The RNA was treated with Turbo DNA–free kit (Applied Biosystems, TX) to remove genomic DNA contamination. A total of 1 g of RNA was reverse transcribed to cDNA using High–Capacity cDNA Reverse Transcription Kit (Applied Biosystems, CA) following the manufacturer’s instructions. After dilution (1/20), 5 μl of cDNA was used for each qRT–PCR in total of 20 μl reaction volume. qRT–PCR was performed using StepOnePlus Real–Time PCR System machine (Applied Biosystems, CA). The qRT–PCR conditions and primer information were described previously (Park et al. 2011). The data from each sample were normalized to the control sample (collected before vaccination) using two housekeeping genes (β–actin and GAPDH) to calculate the relative quantification (RQ) of each cytokine gene transcription at different time points (Park et al. 2011).

Figure 2: Flow cytometric analysis of PBMC in response to live Map stimulation; Data represent the result of FC at 1 wk PV; PBMC were collected at time 0 (A, B, C) or after 6 day culture in RPMI alone (D, E, F) ....
RESULTS
Effect of Vaccination on Blood Circulation and Fecal Shedding of Map
Blood was screened before and after vaccination for the presence of Map by a RT–PCR targeting IS900 sequence due to its high sensitivity compared to a single copy gene (Li et al. 2005; Irenge et al. 2009). Map DNA was detected in blood collected before and at 2 wk PV. Map DNA was below detectable level at 1, 3 and 4 wk PV. For a correct quantification of bacterial shedding, we used a RT–PCR targeting a specific single copy gene of Map, F57, as described by Schonenbrucher et al. (Schonenbrucher et al. 2008). The F57 RT–PCR was less affected by PCR inhibitors present in the DNA samples compared to IS900 RT–PCR (Park et al. 2014). A large number of bacteria were detected in feces before vaccination (1.2 x 104 Map / RT–PCR reaction). Bacterial shedding was reduced after vaccination until 2 wk PV (more than 1 log scale reduction). Map shedding was increased slightly at 3 wk PV, but decreased again at 4 wk PV (Figure 3).
Flow Cytometric Analysis of PBMC Stimulated with Live Map
PBMC stimulation with live Map failed to stimulate PBMC proliferation before vaccination. We tentatively concluded the immune response was anergic and vaccinated the cow to determine if anergy could be reversed. A strong CD4 and CD8 response was detected at one week PV (Figure 2). Proliferation was only observed in the cultures with live Map (Figure 2D and 2G). The specific memory response of CD4 and CD8 T cells to live Map was clearly observed as shown in the panels of CD4 (Figure 2E and 2H) and CD8 (Figure 2F and 2I) gated cells. The proliferating cells expressed CD45R0 and CD25 (data not shown). The proliferative response was similar at 2 and 3 wk PV. Flow cytometry was not conducted at 4 wk PV.
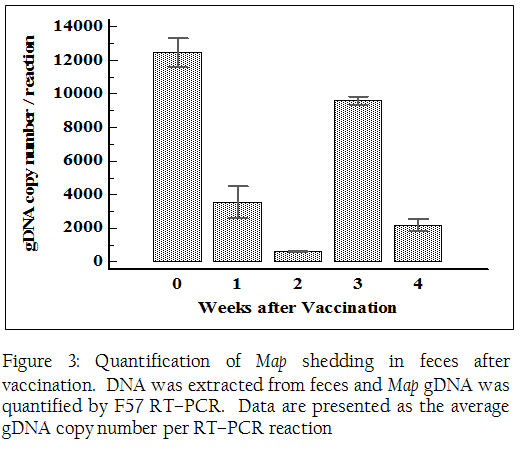
Figure 3: Quantification of Map shedding in feces after vaccination. DNA was extracted from feces and Map gDNA was quantified by F57 RT–PCR. Data are presented as the average gDNA copy number per RT–PCR reaction
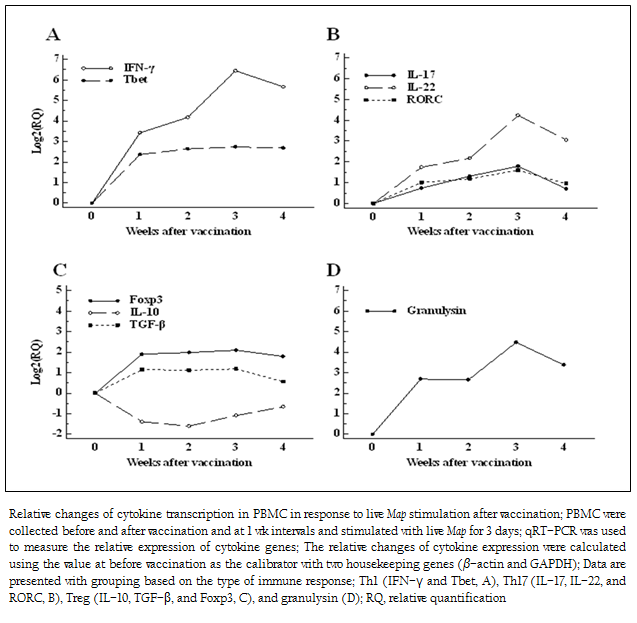
Figure 4: Relative changes of cytokine transcription in PBMC in response to live Map stimulation after vaccination; PBMC were collected before and after vaccination and at 1 wk intervals and stimulated ...
Cytokine Transcription in PBMC Stimulated with Live Map
The relative changes in cytokine expression of PBMC in response to live Map stimulation were compared before and after vaccination. The important signature cytokines for host immune response to Map, such as Th1, Th17, and regulatory T cell (Treg) type cytokines, and a mycobactericidal peptide (granulysin) (Dieli et al. 2001; Gansert et al. 2003), were compared to evaluate the effect of therapeutic vaccination.
The most striking increase in cytokine gene expression after vaccination was IFN–γ. The transcription of IFN–γ gradually increased until 3 wk PV (fold change: 86.4), and then slightly decreased at 4 wk PV (fold change: 50.5). The transcription of Tbet, a transcription factor regulating development of Th1 responses (Zhu et al. 2012), was simultaneously elevated at 1 wk PV (fold change: 5.2) and the level was maintained until 4 wk PV (fold change: 6.4) (Figure 4A). The changes of transcription of Th17 type cytokines (IL–17, IL–22) were lower than Th1 type cytokines. The transcription of IL–17, secreted by Th17 cells, and RORC, a transcription factor for Th17 cells (Ivanov et al. 2006), were just slightly elevated after vaccination (1.6 to 3.4 times for IL–17, and 1.9 to 3.0 times for RORC, respectively). However, the effect of vaccination on inducing another Th17 cytokine, IL–22, was clearly evident. The pattern of transcriptional change of IL–22 was similar to that of IFN–γ. It increased gradually until 3 wk PV (fold change: 18.9), and then slightly dropped at 4 wk PV (Figure 4B). The change of regulatory cytokine gene expression was not significant at any time point. The transcription of TGF–β was slightly up–regulated, while the transcription of IL–10 was down–regulated (Figure 4C). The up–regulation of granulysin gene was also detected after vaccination. The peak increase was observed at 3 wk PV (fold change: 22.3) following the initial increase at 1 wk PV (fold change: 6.3) (Figure 4D).
DISCUSSION
Efforts to characterize the immune response to Map have primarily focused on analysis of the immune response during the early and the latent stages of infection. The cumulative data show that exposure to Map invariably leads to infection and development of an immune response that controls but does not eliminate the pathogen, similar to infection with Mycobacterium tuberculosis (Mtb) (O'Garra et al. 2013). Although studied extensively, it remains unclear which components of the immune response are critical for controlling infection during this latent stage of infection. Cell mediated immunity (CMI) is involved in maintenance of protection and an increase in humoral immunity is associated with disease progression (Stabel 2000). An increase in antibody titer to Map antigens is being used by the dairy industry to predict when animals will develop clinical disease and should be removed from the herd. Few studies have been conducted at the late stage of infection to identify the cellular events associated with a breakdown of protective immunity. The finding that vaccination can transiently reverse immunopathology offers an opportunity to gain insight into how the immune response is being modulated and the components of cellular immunity that are critical for controlling infection (Juste et al. 2009; Singh et al. 2010; Alonso–Hearn et al. 2012). The two most informative studies have shown vaccination can be used at any stage of infection and obtain a therapeutic effect. A field trial showed whole herd vaccination with a commercial vaccine (Silirum, Zoetis), in dairies where Map infection was endemic, reduced bacterial shedding at the herd level and improved milk production (Juste et al. 2009; Alonso–Hearn et al. 2012). A more comprehensive study in goats comparing the therapeutic efficacy of a locally developed killed vaccine (Bison type) and a commercial killed vaccine (Gudair, Spain) showed vaccination significantly reduced shedding of Map, reduced morbidity and mortality, and reversed clinical signs as well as improved physical body condition (Singh et al., 2010). These observations support the use of therapeutic vaccination to increase the duration of the immune response associated with controlling disease progression and show further studies are warranted to find ways to increase the duration of the therapeutic effect. The finding that therapeutic vaccination reactivates the components of the immune response associated with protection also shows that it might be possible to use vaccination at the late stage of infection to identify the components of the immune response critical for maintenance of protection. The present preliminary study was conducted with this in mind and to determine if vaccination with a relA deletion mutant could modulate the immune response similar to killed vaccines. The cow that we obtained to conduct the study was at a very late stage of clinical disease. It had been removed from the herd based on a high titer of antibody to Map, severe diarrhea and weight loss, and reduced productivity. The cow was skin test negative and initial testing for an immune response ex vivo, using our FC assay, showed there was no proliferative response to antigenic stimulation with live Map. qRT–PCR showed expression of cytokine genes associated with inflammatory and suppressive immune responses were not elevated. A proliferative response was observed in CD4 and CD8 cells one week PV. The response was still detectable at three weeks but cell proliferation was not as robust. Although there were fluctuations, the overall Map numbers in blood and feces were reduced following vaccination. Cytokine gene expression was mixed. There was an increase in expression of pro–inflammatory IFN–γ, IL–17, IL–22 genes and associated transcription factors, and granulysin, a bactericidal peptide. There was also an increase in expression of the transcription factor Foxp3 and TGF–β. There was a reduced expression of IL–10.
The promising findings of the current study are that the live attenuated vaccine reversed the immunosuppressive response of the late staged JD cow and induced a CMI response, as measured by CD4 and CD8 proliferation and increased cytokine gene expression. A recent therapeutic vaccination study in clinical JD cattle using a subunit vaccine (heat shock protein 70) showed increased survival and reduced bacterial shedding. However, the effect of the subunit vaccine was limited. It only induced humoral immunity, but not CMI (Santema et al. 2013). The increase in CMI, in addition to the reduced bacterial shedding, after MapΔrelA vaccination in the current study suggests the mutant has a broader therapeutic potential. The findings also indicate that more extensive studies are warranted to determine if the response elicited by vaccination with MapΔrelA increases the length of the therapeutic effect and also identify the components of the immune response associated with the therapeutic effect. Our initial studies have shown expression of multiple activation molecules are up–regulated on PBMC from experimentally and naturally infected animals following stimulation with PPD, soluble antigens, and live Map (Koo et al. 2004; Allen et al. 2009; Allen et al. 2011). Efforts to disrupt the immune response in experimentally infected animals to accelerate disease progression have shown the response is resilient and not readily altered by exposure to high doses of Map, immunosuppressive agents, or by depletion of CD4 cells (Stabel et al. 2009; Allen et al. 2012). Analysis has not revealed the basis for this resilience thus far but has shown the response is complex and involves the interplay of subsets of effector CD4 and CD8 cells secreting combinations of IFN–γ, IL–17, IL–22, and granulysin (Park et al. 2011), subsets that appear to be involved in transient improvement of the immune response at the clinical stage of infection. Comparison of the response mediated by killed or subunit vaccines and MapΔrelA should show whether the live vaccine is more effective in enhancing the therapeutic effect and elucidate which component of the response restores protective immunity.
Taken together, the findings indicate further testing of the mutant as a candidate vaccine should include examining analysis of the cellular events associated with restoring a protective immune response as well as its therapeutic potential.
REFERENCES
Allen, A. J., K. T. Park, G. M. Barrington, K. K. Lahmers, G. S. Abdellrazeq, H. M. Rihan, S. Sreevatsan, C. Davies, M. J. Hamilton and W. C. Davis (2011). "Experimental infection of a bovine model with human isolates of Mycobacterium avium subsp. paratuberculosis." Vet. Immunol. Immunopathol. 141(3–4): 258–266.
http://dx.doi.org/10.1016/j.vetimm.2011.03.014
PMid:21477870 PMCid:PMC3097062
Allen, A. J., K. T. Park, G. M. Barrington, K. K. Lahmers, M. J. Hamilton and W. C. Davis (2009). "Development of a bovine ileal cannulation model to study the immune response and mechanisms of pathogenesis of paratuberculosis." Clin. Vaccine. Immunol. 16(4): 453–463.
http://dx.doi.org/10.1128/CVI.00347-08
PMid:19225077 PMCid:PMC2668272
Allen, A. J., J. R. Stabel, S. Robbe–Austerman, K. T. Park, M. V. Palmer, G. M. Barrington, K. K. Lahmers, M. J. Hamilton and W. C. Davis (2012). "Depletion of CD4 T lymphocytes at the time of infection with M. avium subsp. paratuberculosis does not accelerate disease progression." Vet. Immunol. Immunopathol. 149(3–4): 286–291.
http://dx.doi.org/10.1016/j.vetimm.2012.07.010
PMid:22898538
Alonso–Hearn, M., E. Molina, M. Geijo, P. Vazquez, I. A. Sevilla, J. M. Garrido and R. A. Juste (2012). "Immunization of adult dairy cattle with a new heat–killed vaccine is associated with longer productive life prior to cows being sent to slaughter with suspected paratuberculosis." J. Dairy. Sci. 95(2): 618–629.
http://dx.doi.org/10.3168/jds.2009-2860
PMid:22281327
Bastida, F. and R. A. Juste (2011). "Paratuberculosis control: a review with a focus on vaccination." J Immune Based Ther Vaccines 9: 8.
http://dx.doi.org/10.1186/1476-8518-9-8
PMid:22035107 PMCid:PMC3222599
Bull, T. J., S. C. Gilbert, S. Sridhar, R. Linedale, N. Dierkes, K. Sidi–Boumedine and J. Hermon–Taylor (2007). "A Novel Multi–Antigen Virally Vectored Vaccine against Mycobacterium avium subspecies paratuberculosis." PLoS. ONE. 2(11): e1229.
http://dx.doi.org/10.1371/journal.pone.0001229
PMid:18043737 PMCid:PMC2082073
Burke, D. S. (1993). "Of postulates and peccadilloes: Robert Koch and vaccine (tuberculin) therapy for tuberculosis." Vaccine. 11(8): 795–804.
http://dx.doi.org/10.1016/0264-410X(93)90354-Z
Dieli, F., M. Troye–Blomberg, J. Ivanyi, J. J. Fournie, A. M. Krensky, M. Bonneville, M. A. Peyrat, N. Caccamo, G. Sireci and A. Salerno (2001). "Granulysin–dependent killing of intracellular and extracellular Mycobacterium tuberculosis by Vgamma9/Vdelta2 T lymphocytes." J. Infect. Dis. 184(8): 1082–1085.
http://dx.doi.org/10.1086/323600
PMid:11574927
Faisal, S. M., J. W. Chen, F. Yan, T. T. Chen, N. M. Useh, W. Yan, S. Guo, S. J. Wang, A. L. Glaser, S. P. McDonough, B. Singh, W. C. Davis, B. L. Akey and Y. F. Chang (2013). "Evaluation of a Mycobacterium avium subsp. paratuberculosis leuD mutant as a vaccine candidate against challenge in a caprine model." Clin. Vaccine. Immunol. 20(4): 572–581.
http://dx.doi.org/10.1128/CVI.00653-12
PMid:23408524 PMCid:PMC3623397
Gansert, J. L., V. Kiessler, M. Engele, F. Wittke, M. Rollinghoff, A. M. Krensky, S. A. Porcelli, R. L. Modlin and S. Stenger (2003). "Human NKT cells express granulysin and exhibit antimycobacterial activity." J. Immunol. 170(6): 3154–3161.
http://dx.doi.org/10.4049/jimmunol.170.6.3154
PMid:12626573
Irenge, L. M., K. Walravens, M. Govaerts, J. Godfroid, V. Rosseels, K. Huygen and J. L. Gala (2009). "Development and validation of a triplex real–time PCR for rapid detection and specific identification of M. avium subsp. paratuberculosis in faecal samples." Vet. Microbiol. 136(1–2): 166–172.
http://dx.doi.org/10.1016/j.vetmic.2008.09.087
PMid:19095382
Ivanov, II, B. S. McKenzie, L. Zhou, C. E. Tadokoro, A. Lepelley, J. J. Lafaille, D. J. Cua and D. R. Littman (2006). "The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL–17+ T helper cells." Cell. 126(6): 1121–1133.
http://dx.doi.org/10.1016/j.cell.2006.07.035
PMid:16990136
Juste, R. A., M. Alonso–Hearn, E. Molina, M. Geijo, P. Vazquez, I. A. Sevilla and J. M. Garrido (2009). "Significant reduction in bacterial shedding and improvement in milk production in dairy farms after the use of a new inactivated paratuberculosis vaccine in a field trial." BMC. Res. Notes. 2: 233.
http://dx.doi.org/10.1186/1756-0500-2-233
PMid:19930604 PMCid:PMC2788577
Kathaperumal, K., V. Kumanan, S. McDonough, L. H. Chen, S. U. Park, M. A. Moreira, B. Akey, J. Huntley, C. F. Chang and Y. F. Chang (2009). "Evaluation of immune responses and protective efficacy in a goat model following immunization with a coctail of recombinant antigens and a polyprotein of Mycobacterium avium subsp. paratuberculosis." Vaccine. 27(1): 123–135.
http://dx.doi.org/10.1016/j.vaccine.2008.10.019
PMid:18955101
Koo, H. C., Y. H. Park, M. J. Hamilton, G. M. Barrington, C. J. Davies, J. B. Kim, J. L. Dahl, W. R. Waters and W. C. Davis (2004). "Analysis of the immune response to Mycobacterium avium subsp. paratuberculosis in experimentally infected calves." Infect. Immun. 72(12): 6870–6883.
http://dx.doi.org/10.1128/IAI.72.12.6870-6883.2004
PMid:15557608 PMCid:PMC529129
Li, L., J. P. Bannantine, Q. Zhang, A. Amonsin, B. J. May, D. Alt, N. Banerji, S. Kanjilal and V. Kapur (2005). "The complete genome sequence of Mycobacterium avium subspecies paratuberculosis." Proc. Natl. Acad. Sci. USA. 102(35): 12344–12349.
http://dx.doi.org/10.1073/pnas.0505662102
PMid:16116077 PMCid:PMC1194940
O'Garra, A., P. S. Redford, F. W. McNab, C. I. Bloom, R. J. Wilkinson and M. P. Berry (2013). "The immune response in tuberculosis." Annu. Rev. Immunol. 31: 475–527.
http://dx.doi.org/10.1146/annurev-immunol-032712-095939
PMid:23516984
Park, K. T., A. J. Allen, J. P. Bannantine, K. S. Seo, M. J. Hamilton, G. S. Abdellrazeq, H. M. Rihan, A. Grimm and W. C. Davis (2011). "Evaluation of two mutants of Mycobacterium avium subsp. paratuberculosis as candidates for a live attenuated vaccine for Johne's disease." Vaccine. 29(29–30): 4709–4719.
http://dx.doi.org/10.1016/j.vaccine.2011.04.090
PMid:21565243 PMCid:PMC3114198
Park, K. T., A. J. Allen and W. C. Davis (2014). "Development of a novel DNA extraction method for identification and quantification of Mycobacterium avium subsp. paratuberculosis from tissue samples by real–time PCR." J. Microbiol. Methods. doi: 10.1016/j.mimet.2014.02.003.
http://dx.doi.org/10.1016/j.mimet.2014.02.003
Park, K. T., J. L. Dahl, J. P. Bannantine, R. G. Barletta, J. Ahn, A. J. Allen, M. J. Hamilton and W. C. Davis (2008). "Demonstration of allelic exchange in the slow–growing bacterium Mycobacterium avium subsp. paratuberculosis, and generation of mutants with deletions at the pknG, relA, and lsr2 loci." Appl. Environ. Microbiol. 74(6): 1687–1695.
http://dx.doi.org/10.1128/AEM.01208-07
PMid:18192416 PMCid:PMC2268294
Rosseels, V. and K. Huygen (2008). "Vaccination against paratuberculosis." Expert. Rev. Vaccines. 7(6): 817–832.
http://dx.doi.org/10.1586/14760584.7.6.817
PMid:18665779
Santema, W., V. Rutten, R. Segers, J. Poot, S. Hensen, H. Heesterbeek and A. Koets (2013). "Postexposure subunit vaccination against chronic enteric mycobacterial infection in a natural host." Infect. Immun. 81(6): 1990–1995.
http://dx.doi.org/10.1128/IAI.01121-12
PMid:23509147 PMCid:PMC3676026
Scandurra, G. M., G. W. de Lisle, S. M. Cavaignac, M. Young, R. P. Kawakami and D. M. Collins (2010). "Assessment of Live Candidate Vaccines for Paratuberculosis in Animal Models and Macrophages." Infect. Immun. 78(3): 1383–1389.
http://dx.doi.org/10.1128/IAI.01020-09
PMid:20038535 PMCid:PMC2825941
Schonenbrucher, H., A. Abdulmawjood, K. Failing and M. Bulte (2008). "New triplex real–time PCR assay for detection of Mycobacterium avium subsp. paratuberculosis in bovine feces." Appl. Environ. Microbiol. 74(9): 2751–2758.
http://dx.doi.org/10.1128/AEM.02534-07
PMid:18326682 PMCid:PMC2394907
Singh, S. V., P. K. Singh, A. V. Singh, J. S. Sohal and M. C. Sharma (2010). "Therapeutic Effects of a New "Indigenous Vaccine" Developed Using Novel Native "Indian Bison Type" Genotype of Mycobacterium avium Subspecies paratuberculosis for the Control of Clinical Johne's Disease in Naturally Infected Goatherds in India." Vet. Med. Int. 2010: 351846.
http://dx.doi.org/10.4061/2010/351846
PMid:20445782 PMCid:PMC2860218
Stabel, J. R. (2000). "Transitions in immune responses to Mycobacterium paratuberculosis." Vet. Microbiol. 77(3–4): 465–473.
http://dx.doi.org/10.1016/S0378-1135(00)00331-X
Stabel, J. R., A. Barnhill, J. P. Bannantine, Y. F. Chang and M. A. Osman (2012). "Evaluation of protection in a mouse model after vaccination with Mycobacterium avium subsp. paratuberculois protein cocktails." Vaccine. 31(1): 127–134.
http://dx.doi.org/10.1016/j.vaccine.2012.10.090
PMid:23137840
Stabel, J. R., M. V. Palmer, B. Harris, B. Plattner, J. Hostetter and S. Robbe–Austerman (2009). "Pathogenesis of Mycobacterium avium subsp. paratuberculosis in neonatal calves after oral or intraperitoneal experimental infection." Vet. Microbiol. 136(3–4): 306–313.
http://dx.doi.org/10.1016/j.vetmic.2008.11.025
PMid:19135813
Zhu, J., D. Jankovic, A. J. Oler, G. Wei, S. Sharma, G. Hu, L. Guo, R. Yagi, H. Yamane, G. Punkosdy, L. Feigenbaum, K. Zhao and W. E. Paul (2012). "The transcription factor T–bet is induced by multiple pathways and prevents an endogenous Th2 cell program during Th1 cell responses." Immunity. 37(4): 660–673.
http://dx.doi.org/10.1016/j.immuni.2012.09.007
PMid:23041064 PMCid:PMC3717271