Advances in Animal and Veterinary Sciences
Research Article
Effect of Leptin Gene Polymorphism on Reproductive Efficiency in Awassi Ewes
Laith Sofian Younis1*, Hayder Abd Alkarem Al-Mutar2, Ali Aziz Abid1
1College of Veterinary Medicine, Tikrit University, Iraq; 2College of Veterinary Medicine, Baghdad University, Iraq.
Abstract | LEPtin (LEP) is a hormone that strongly associate with nutritional state, glucose homeostasis and reproduction. Study performed to identify the linkage between LEP polymorphism and reproductive efficiency such as Seasonality and litter size. Forty mature non-pregnant Awassi ewes were utilized between 1st July/2017 to 1st May/2018 in Salah Aldin province/Iraq. Twenty ewes were demonstrated estrus heat at August/2017which considered Seasonal group, and the others showed estrus signs at April/2018, which considered Non-Seasonal group.Genomic DNA was extracted from blood specimens and four primers were utilized to amplify exon II, intron II (fragment 1) and exon III (fragment 1) of LEP geneby polymerase chain reaction (PCR). Polymorphisms were revealed via sequencing and compared with the sequencing of the ovine LEP gene in NCBI. One single nucleotide polymorphism (SNP) A(99)R was detected in intron II and three SNPs G(425)R, T(541)K and G(587)R were in exon III. Two genotypes of each SNP were observed with higher significant differences (P<0.01) between frequencies 47.50 and 52.50 for AA and AG of A(99)R, 42.50 and 57.50 for GG and GA of G(425)R, 55.00 and 45.00 for TT and TC of T(541)K, lastly, 37.50, 62.50 for GG and GA of G(587)R. The finding demonstrated AG genotypic frequency of Non-Seasonal ewes (55.00) was significantly increased (P<0.05) than AA (45.00) for A(99)R SNPs. The mutant GA, TC and GA genotypic frequencies (80.00, 40.00 and 85.00) were recorded higher significant increased (P<0.01) than wild genotype GG, TT and GG (20.00, 60.00 and 15.00) for G(425)R, T(541)K and G(587)R SNPs in Non-Seasonal group. Higher significant increased (P<0.01) were observed in GG, TT and GG genotypic frequencies (65.00, 70.00 and 60.00) than GA, TC and GA (35.00, 30.00 and 40.00) for G(425)R, T(541)K and G(587)R SNPs respectively in Seasonal group, while non-significant differences were observed between AA and AG (50.00 for each) genotypic frequency for A(99)R SNPs of Seasonal group. Non-significant differences in litter size were recorded between GA (1.39) and GG (1.29) for G(425)R and between GG (1.4) and GA (1.32) for G(587)R, while significant differences (P<0.05) were observed between AG (1.26) and AA (1.42) and between TT (1.18) and TC (1.55) for A(99)R and T(541)K respectively. In Awassi breed, exon III polymorphisms of LEP gene have an expected effect on the Seasonality and the wild genotypes find majorly in Seasonal ewes. Intron II and exon III polymorphisms (2 SNPs) caused an increment in litter size.
Keywords | LEPtin, Litter Size, Polymorphism, Reproduction, Seasonality.
Editor | Kuldeep Dhama, Indian Veterinary Research Institute, Uttar Pradesh, India.
Received | August 07, 2018; Accepted | October 04, 2018; Published | November 02, 2018
*Correspondence | Laith Sofian Younis, College of Veterinary Medicine, Tikrit University, Iraq; Email: [email protected]
Citation | Younis LS, Al-Mutar HAA, Abid AA (2019). Effect of leptin gene polymorphism on reproductive efficiency in awassi ewes. Adv. Anim. Vet. Sci. 7(1): 17-23.
DOI | http://dx.doi.org/10.17582/journal.aavs/2019/7.1.17.23
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2019 Younis et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
LEPtin (LEP) is a protein release mostly from adipose tissue (the major source of LEP) (Houseknecht et al., 1998). It consists of 146 amino acids with molecular weight 16000 Dalton (Zhang et al., 1994). Chromosome four is containing the ovine LEP gene (Moravcikova et al., 2012). It’s included two introns and three exons, the last two exons are involved in LEP protein synthesis (167 amino acids) which is undergo cleavage of 21 amino acid (signal peptide) (De La et al., 1996; Zhang et al., 1994).
LEPtin receptors are categorized as class one cytokine receptors because of bilaterally symmetrical with a structure of IL-6 receptors (Houseknecht and Portocarrero, 1998). Because the LEP mRNA receptors are extensively express in arcuate nuclei, ventromedial hypothalamus and adenohypophysis of ewes, LEP act directly on brain and pituitary levels to promote and coordinate gonadotropin (FSH/LH) secretion (Dyer et al., 1997). The LEP infusion cause restore Kiss1 mRNA expression of kisspeptin neurons in an Arcuate nucleus and Preoptic area for poorly nourished ewes, and kisspeptin regulate brain control of reproduction by a reduction in proopiomelanocortin (POMC) and raised neuropeptide Y (NPY) gene expression (Backholer et al., 2010).
In sheep; LEP blood circulating level is correlate with nutritional levels (Marie et al., 2001), feeding value (Blache et al., 2000) and condition of fat mass (Delavaud et al., 2000). The main role of LEP is regulating GnRH and prevents reduction of pulsatile LH throughout fasting (Nagatani et al., 1998) because GnRH neurons affect directly by LEP (Sullivan and Meonter, 2004). A severe feed restriction for 48 h caused an apparent decline in LEP secretion concurrent with LH drooping in cow (Amstalden et al., 2000) and ewes (Henry et al., 2001; Morrison et al., 2001). Continual feed deprivation for seven days gives rise to decline both of serum LEP and LH ovariectomized (OVX) young sow (Whisnant and Harrell, 2002).
In fasted cows; LEP treatments stimulate adenohypophyseal LH secretion mediated by basal GnRH (Amstalden et al., 2003). Furthermore, LEP blocks the pulsating reduction of LH and promote GnRH secretion in heifers (Maciel et al., 2004). Moreover, sexual immaturity is correlating with low LEP levels (El-Eshmawy and Aal, 2010).
In ewes, LH secretion stimulated by intra cerebro ventricular (ICV) LEP injection in feed restricted OVX ewe (Henry et al., 2001). In spite of the fact that chronic ICV administration of LEP failed to stimulate LH secretion in well-fed ewes (Henry et al., 1999), the LEP injection caused rise of LH blood concentration and non-significantly enhance blood FSH level in feed deprivation; these facts give an indication about role of LEP as a metabolic signal on GnRH-LH/FSH axis in feed-limited ewes (Towhidi et al., 2007).
Several genetic SNPs contributed with Seasonality in sheep; aryl alkyl amine-N-acetyl-transferase gene (AA-NAT) and melatonin receptor 1A gene polymorphism correlated with ewes that breeding out of season (Giantsis et al., 2016; Hatif and Younis, 2018). As well as, GDF9 gene SNPs in Araucana creole sheep breed linked with litter size (Bravo et al., 2016). In sow, LEP receptor gene polymorphism record increment in litter size for Yorkshire and Duroc breed (Chen et al., 2004a).
Numerous studies pointed out about the role of LEP gene polymorphisms and growth parameters in different domestic animals; dairy cattle production quality (Nkrumah et al., 2004), sheep muscles growth and meat features (Boucher et al., 2006), body weight and growth traits (Shojaei et al., 2011).
Because the role of LEP on GnRH/LH and since the available information regarding the effect of polymorphism,of LEP gene on sheep reproduction aspect and (hypothalamic-pituitary-gonadal axis) is insufficient, therefore, the existing study was carried out to identify polymorphisms in ovine LEP gene3 and their possible association with the time of estrus cycle and litter size in Iraqi Awassi ewes.
Material and methods
Animals Management and Samples Collection
Three ml blood specimens were, collected randomly during April from forty mature multiparous Awassi ewes with average age three years in Salah Aldin province. The animals were disconnected into two categories depending on breeding season; about 20 of ewes showed estrus at august/2017 (Seasonal ewes) (group 1) and the rest were demonstrated estrus cycle at April /2018 (Non-Seasonal ewes) (group 2). At all time; animals were housed in one flock with a breeding ram in the animal house of veterinary college/Tikrit university; that locate in latitude 34 and longitude 43. The blood samples were septically aspirated from vena puncture of jugular vein into vitamin K containing collection tube (APTACA/Italy) and stored at -20°C for DNA extraction, amplification and sequencing of exon II and III of LEP gene.
Genomic DNA was extracted by utilized G-spin Kit (Intron/ Korea) according to manufacturer’s protocol. Four Primers were designed based on genomic sequences of sheep (Genbank, AF310264, and AY831682) and (GenBank, U84247, and AY911719) according to Boucher et al. (2006). The sequence of primer 1 forward: 5´-CGCAAGGTCCAGGATGACACC-3´; and primer 1 reverse: 5´-GTCTGGGAGGGAGGAGAGTGA-3´ that’s amplified part of exon II and intron II (fragment 1), the sequence of primer 2 forward: 5’CTCTTGATGTCCCCTTCCTC-3’ and primer 2 reverse: 5’ TGGTCCTTCGAGATCCATTC-3’ that’s designed to amplified all exon III and part of the 3′ UTR of the gene (fragment 2) (Mahmoud et al., 2014). The total volume of reactions were 25 μl; that containing 2 ul of genomic DNA, 1 μl of each primer (10 pM), PCR Master Mix Kit (Intron/ Korea) 12.5 ul, and nuclease-free water 8.5 ul in 30 cycles for fragment 1 and 2 (initial denaturation: 95°C for 5 min, denaturation: 95°C for 30s, annealing: 60°C for 30s for fragment 1 and 54 °C for 30 sec for fragment 2, extension: 72°C for 30s and final extension 72°C for 7 min).
Table 1: The SNPs location, number, nucleotide and changes of amino acidsfor Awassi ewes
| Location Of SNP | SNP Number | Nucleotide Change | Amino Acid Change & Number | Predicted Effect | Type of Mutation | |
| 1 |
A(99)R (Intron II) |
Rs408463464 | TAG>TAG& TAA | |||
| 2 |
G(425)R (Exon III) |
Rs409584889 | CGG> CAG& CGG | Arginine > Glutamine & Arginine (142) | Transition | Missense |
| 3 |
T(541)K (Exon III) |
Rs420693815 | TTG> GTG& TTG | Leucine>Valine & Leucine(181) | Trasversion | Missense |
| 4 |
G(587)R (Exon III) |
Rs428185456 | CGG> CAG& CGG | Arginine > Glutamine & Arginine (196) | Transition | Missense |
Table 2: The observed genotypic and allele frequencies of LEP gene for Awassi ewes
| No | Locus | Genotypes | Observed Genotypes | Genotypic Frequency % | Allele Frequency | Chi Square | |
|
1 |
A(99)R |
AA | 19 | 47.50 | A | 73.75 | 11.72 ** |
| AG | 21 | 52.50 | G | 26.25 | |||
|
2 |
G(425)R | GG | 17 | 42.50 | G | 71.25 | 11.55 ** |
| GA | 23 | 57.50 | A | 28.75 | |||
|
3 |
T(541)K | TT | 22 | 55.00 | T | 77.5 | 12.42 ** |
| TC | 18 | 45.00 | C | 22.5 | |||
|
4 |
G(587)R | GG | 15 | 37.50 | G | 68.75 | 10.09 ** |
| GA | 25 | 62.50 | A |
31.25 |
|||
** (P<0.01).
The DNA bands were detected by placing ethidium bromide staining gel electrophoresis in Trans illuminator (Vilberlourmat/ France).
Amplicons of LEP gene were sequenced separately by Macrogen Corporation/ Korea. The LEP sequences were edited, and aligned, by utilizing (BioEdit software). The samples homology done by applied BLAST option in NCBI GenBank database and Bioedit program.
Statistical Analysis
Statistical Analysis System”- SAS (2012) program was utilized to monitor the effect of different factors in study parameters. T-test and Chi-square test were usedto significant compared between means and between percentages respectively in this study.
Results and discussion
PCR Amplification
Two primers that amplified particular regions (part of exonII and part of intronII) of LEP gene of Awassi ewes. The PCR amplified size was 260 bp. In addition to that, another twoprimers were amplified of exonIII and part of 3′ UTR and the uniform fragments with size 1090 bp appeared after electrophoresed in 1% agarose gel (Figure 1).
Sequencing and Genetic Variability
The sequencing result revealed that one SNP in Intron II A(99)R and three SNPs in exon III (G(425)R, T(541)K
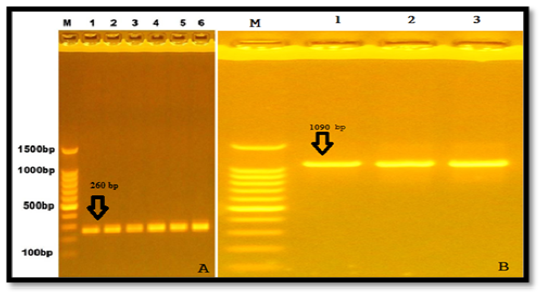
Figure 1: Results of the presence of LEP gene of samples were fractionated on 1% agarose gel electrophoresis stained with ethidium bromide Lane1:100bp DNA marker A. Exon II& intron II, B. Exon III.
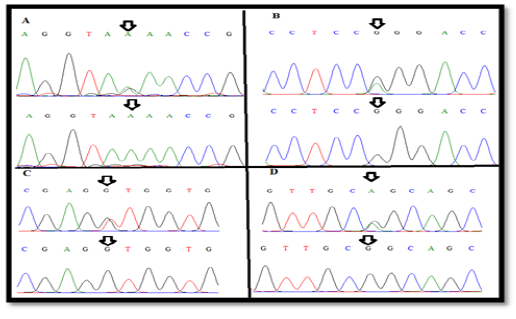
Figure 2: The wild-type and new variant (A) A(99)R , (B) G(425)R (C), (C) T(541)K and (D) G(587)R of exonII, intronII and exonIII of LEP gene
Table 3: Chi- square of genotypic distribution of LEP gene inNon-Seasonal and Seasonal ewes.
| No | Locus | Genotypes | Genotypic Frequency for Non-Seasonal Ewes | Chi- Square | Genotypic Frequency for Seasonal Ewes | Chi- Square |
|
1 |
A(99)R |
AA |
9 (45%) |
4.57 * |
10 (50%) |
0.00 NS |
|
AG |
11 (55%) |
10 (50%) |
||||
|
2 |
G(425)R |
GG |
4 (20%) |
13.86 ** |
13 (65%) |
9.87 ** |
|
GA |
16 (80%) |
7 (35%) |
||||
|
3 |
T(541)K |
TT |
8 (40%) |
7.25 ** |
14 (70%) |
10.33 ** |
|
TC |
12 (60%) |
6 (30%) |
||||
|
4 |
G(587)R |
GG |
3 (15%) |
13 .46 ** |
12 (60%) |
7.25 ** |
|
GA |
17 (85%) |
8 (40%) |
* (P<0.05), ** (P<0.01).
Table 4: T- test of the litter size for the LEP genotypes of Awassi ewes
| Locus | Genotypes | Ewe Number | Lambs Number | Litter Size | T- Test | |
| 1 | A(99)R | AA | 19 | 24 | 1.26 | 0.128 * |
| AG | 21 | 30 | 1.42 | |||
| 2 | G(425)R | GG | 17 | 22 | 1.29 | 0.115 NS |
| GA | 23 | 32 | 1.39 | |||
| 3 | T(541)K | TT | 22 | 26 | 1.18 | 0.196 * |
| TC | 18 | 28 | 1.55 | |||
| 4 |
G(587)R
|
GG | 15 | 21 | 1.4 | 0.107 NS |
| GA | 25 | 33 |
1.32 |
* (P<0.05).
and G(587)R) of LEP gene in Awassi ewes (Figure 2). All these SNPs were recorded in NCBI and Ensembl gene browser (rs408463464, rs409584889, rs420693815 and rs428185456 respectively). The last three SNPs of coding region are missense mutation that changed amino acid to another; Arginine > Glutamine, Leucine> Valine and Arginine > Glutamine in position 142, 181 and 196 respectively (Table 1).
Significant differences were recorded between the genetic variability of LEP gene. Higher significant variation (P<0.001) were recorded between the (AA and AG), (GG and GA), (TT and TC) and (GG and GA) genotypes of A(99)R, G(425)R, T(541)K and G(587)R locus respectively, while GG, AA, CC and AA genotypes didn’t noticed in these populations of the same locus (Table 2).These amino acids changes are impacted in LEP hormone function.These are in agreement with the Zhou et al. (2009) finding, which hypothesized that diversity of LEP gene might have an influence on LEP activity and function.
Correlation between Genotypes and Breeding Season in Awassi Sheep
The genotypic distributions of LEP gene in the two animal groups were recorded. In Non-Seasonal Awassi ewes group, higher significantly increased (P<0.001) were showed in mutant genotypes;GA, TC and GA as compared with the wild genotypes; GG, TT and GG of G(425)R, T(541)K and G(587)R locus, also AG genotype of A(99)R locus recorded significantly increased (P<0.05) when compared with AA genotype . On the other hand, higher significantly increased (P<0.001) were recorded in wild genotypes; GG, TT and GG in comparison with other genotypes (GA, TC and GA) of G(425)R, T(541)K and G(587)R locus respectively, while non-significant differences were recorded between AG and AA genotypes for A(99)R locus for Seasonal Awassi ewes group (Table 3).
This finding leads to speculate that the mutations has affected on the final form of LEP protein and caused increasing its function in ewes that breed off season. Only a few studies have been performed, to detect the relationship between polymorphism, of LEP,gene and ovine reproductive performance; Bravo et al. (2016) find a relationship between LEP gene polymorphism and litter size. When compared the sequence of exon III for the Najdi breed (ID: KF922846.1), the GG, TT and GG genotypic frequency in (G(425)R, T(541)K and G(587)R were present, while AG, TC and GA genotypes (mutant alleles) of the same locus were absent, the Najdi breed has shorter lambing interval compared with the Awassi breed (330 vs 286 days) (Abdelqader et al., 2012), that means the Non-Seasonal Awassi group has a longer breeding season than Najdi breed.
The LEP hormone is a metabolic factor that has an important role in up-regulation GnRH and LH output by an effect on kisspeptin, POMC and NPY expression in the arcuate nucleus and preoptic area of ewe’s hypothalamus (LEP receptors are expressed in kisspeptin cells) (Backholer et al., 2010). Therefore, the SNPs in LEP gene were strongly contributed to reproduction by an effect on kisspeptin secretion because the missense mutation make structural changes in protein conformation and that may enhance LEP protein function. These observations come constantly with several studies that revealed the polymorphisms in exon III of LEP gene were contributed with body weight and growth rate in Kermani sheep (Shojaei et al., 2011) and in Baluchi sheep (Tahmoorespur et al., 2010). These facts find out that LEP gene polymorphism have a positive influence of reproduction through its effect on fat deposition because the adipose tissue is the central source of LEP.This hypothesis was accord with Buchanan et al. (2002) study; which showed that LEP gene polymorphism contributed to increased expression of mRNA LEP gene. These event lead to in plasma LEP elevation (Liefers et al., 2003), and when LEP level increased, the LH is also increased. These argue was agreement with Henry et al. (2001), who mentioned that the LEP cerebral injection elevated of LH secretion. Also, GnRH secretion is indirectly potentiated and regulated by LEP because itacts as a metabolic factor (Burcelin et al., 2003; Quennell et al., 2009).
Correlation between genotypes, and litter, size in Awassi sheep
The results, of present study indicates that LEP gene contribute to the ovulation rate and subsequently litter size. The finding appeared that LEP AG and TC genotypes had significantly raised (P<0.05) in litter size when compared with AA and TT genotypes ofA(99)R and T(541)K respectively. Non-significant differences,were noticed between each genotype of G(425)R and G(587)R (Table 4).
Litter size is correlated with ovulation rate, the finding of present study showed variance between SNPs and genotypes; non-significant differences in litter size were noticed between the genotypes of each (G(425)R and G(587)R, while the genotypes of in locus in A(99)R intron II and the genotypes of T(541)K locus inexon III were recorded significant differences that reflect the mediated effect of LEP gene polymorphism on litter size, these outcomes have not been described previously in sheep. In spite the fact that mRNA of LEP receptor,not expressed, in ovine granulosa and the ca cells (Spicer, 2001), but in this study the polymorphisms recorded increase in ovulation rate. These results accord with Chen et al. (2004b) outcomes, which find that the litter size was increased significantly in mutant allele of porcine,LEP gene. As mentioned above; LEP increases GnRH and LH in ewes which reflected positively on ovulation rate (litter size). Additionally, according to Lagonigro et al. (2003), theincrease fat deposition and feed intake occurs because LEP gene polymorphism andthat explain the effect of polymorphism on enhancement the litter size. This speculation was corresponded with Kara et al. (2010), who mentioned that improving feed intake had a positive effect on reproduction and litter size Awassi ewes.
Taking into,consideration,the vast range of physiological LEP impact, it is expected that several phenotypic features have been detected with LEP gene SNPs. As a conclusion, heterozygote LEP gene polymorphisms of exon III were related with Awassi ewes that breeding out of season, while wild genotypic frequencies were higher in Seasonal Awassi ewes. Litter size was correlated with two SNPs, in intron II and exon III.
Acknowledgements
The authors thank the staff of laboratory of Biotechnology Research Center-Al-Nahrian University/Baghdad and laboratory of veterinary college/Tikrit University. Also special thanks to employers of animal house/veterinary college/Tikrit University.
Conflict of interest
There is no conflict of interest.
authors contribution
Laith Sofian Younis supervised the project. Both Hayder Alkarem Al-Mutar and Laith Sofian Younis designed and performed the experiments of genotyping, while Ali Aziz Abd wrote the manuscript, inconsultation with Hayder Alkarem Al-Mutar and Laith Sofian Younis. Also Ali Aziz Abd analyzed the data.
References






