Advances in Animal and Veterinary Sciences
Research Article
Microbiological and Molecular Detection of Burkholderia vietnamiensis from Nasal Swabs of Small Ruminants and Soil in Nueva Ecija, Philippines
Gabriel Alexis SP. Tubalinal1,3, Danica C. Gregorio2, Jerwin R. Undan2, Claro N. Mingala1,3,4*
1College of Veterinary Science and Medicine, Central Luzon State University, Science City of Muñoz 3120, Nueva Ecija, Philippines; 2Department of Biology, College of Arts and Sciences, Central Luzon State University, Science City of Muñoz 3120, Nueva Ecija, Philippines; 3Biosafety and Environment Section, Philippine Carabao Center, Maharlika Highway, Science City of Munoz 3120, Nueva Ecija, Philippines; 4Department of Animal Science, College of Agriculture, Central Luzon State University, Science City of Muñoz 3120, Nueva Ecija, Philippines.
Abstract | The genus Burkholderia is a diverse group of morphologically, metabolically, and ecologically gram-negative bacteria. The extent and burden of Burkholderia in the Philippines is still undetermined due to the lack of extensive research on this bacterium and the role and relationship of livestock with the bacterium. This study was conducted to detect Burkholderia sp. in the province of Nueva Ecija, the Philippines. Isolation and detection of Burkholderia sp. were demonstrated and validated by microbiological and molecular assays. Twenty-five (25) nasal swabs from goat and sheep and ten (10) soil samples were randomly collected in the area of Nueva Ecija and subjected to bacterial culture. Detection of the bacteria molecularly conducted through the amplification of the 16s rDNA and Tat-domain protein. In addition, amplified PCR products of the Tat-domain protein gene were subjected to sequencing. Results showed that one (1) isolate from goat nasal swab was positive for both the 16s rDNA and the Tat-domain protein of Burkholderia sp. Sequencing revealed 97.86% homology to Burkholderia vietnamiensis strain G4. Furthermore, sequence analysis showed 95%-97% homology to different strains of B. vietnamiensis. This is the first report of B. vietnamiensis isolated in a nasal swab of goat in the Philippines using microbiological and molecular techniques. Furthermore, high-throughput sequence analysis is recommended to further analyze the B. vietnamiensis isolate in the Philippines.
Keywords | Burkholderia vietnamiensis; 16s rDNA; Tat-domain protein; Philippines
Received | February 11, 2021; Accepted | February 25, 2021; Published | March 30, 2021
*Correspondence | Claro N. Mingala, Philippine Carabao Center, Maharlika Highway, Science City of Munoz 3120, Nueva Ecija, Philippines; Email: [email protected]
Citation | Tubalinal GASP, Gregorio DC, Undan JR, Mingala CN (2021). Microbiological and Molecular Detection of Burkholderia vietnamiensis from Nasal Swabs of Small Ruminants and Soil in Nueva Ecija, Philippines. Adv. Anim. Vet. Sci. 9(5): 766-772.
DOI | http://dx.doi.org/10.17582/journal.aavs/2021/9.5.766.772
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2021 Mingala et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
The genus Burkholderia is a very diverse group of morphologically, metabolically, and ecologically gram-negative bacteria. Burkholderia sp. is abundant in the environment with an extensive variety of niches ranging from soil to human and animal respiratory tract (Sawana et al., 2014). Several strains of Burkholderia are reported to be able to fix atmospheric nitrogen that can be used by certain crops to enhance crop yields. However, the use of Burkholderia sp. as biofertilizer is still in contention as some species that fix nitrogen may also cause disease in immunocompromised humans (Sandanakirouchenane et al., 2017).
In the field of medicine, three of the most important species of Burkholderia include B. cepacia complex (BCC), B. pseudomallei and B. mallei. BCC, the term used for the 17 closely related Burkholderia sp., is responsible for a widespread and deadly respiratory disease like cystic fibrosis in immunocompromised individuals (Biddick et al., 2003; Hauser et al., 2011; Sawana et al., 2014). B. pseudomallei is the causative agent of melioidosis, a potentially deadly infectious disease that may account for up to 20% of community-acquired septicemias in tropical regions (Limmathurotsakul and Peacock, 2011; Sandanakirouchenane et al., 2017). Lastly, B. mallei, the causative agent of glanders, cause febrile disease of solipeds (Van Zandt et al., 2013; Sandanakirouchenane et al., 2017). Furthermore, this Burkholderia species are zoonotic and can be used as an agent for bioterrorism.
The status and burden of these diseases in the Philippines are unknown. Furthermore, the causative agent of these diseases, especially B. pseudomallei, were detected only in human clinical cases and some research animals in the Philippines (Whetmore and Gochenour, 1956; Dance et al., 1992; San Martin et al., 2018) and was never reported in livestock. The study focused on microbiological and molecular identification of Burkholderia sp. isolated from nasal swabs of small ruminants and soil in Nueva Ecija, Philippines.
MATERIALS AND METHODS
Collection and Preparation of Samples
Nasal swab samples from 25 small ruminants (15 goats and 10 sheep), regardless of health status, were collected using a sterile cotton swab placed in sterile plastic tubes to prevent contamination and stored in a cooler with ice packs during transport. Furthermore, eight (8) soil samples within the area of the animals were raised were also collected using the simplified standard operating procedure of soil sampling by Wuthiekanun and Dance (2012) with minor modifications. Within the sampling area (50 x 50 m2), a fixed-interval sampling grid was used to collect samples per field, 5 m apart. For each sample, about 100-g soil was collected from a depth of 30 cm.
At the laboratory, the soil samples were prepared using the procedure of Lau et al. (2014) for bacterial enrichment. Ten (10) g of soil and 25 ml of distilled water were placed on tubes and shaken vigorously to dissolve the soil. The tubes were left to stand for 24 h, and 1 µL was collected from the uppermost layers for culture.
Bacterial Culture
The bacterial culture method was performed using a blood agar base (Hi-Media, India) with 5% sheep blood and gentamicin. The preparation of the blood agar was adapted from Buxton (2005) with minor modifications. Ten (10) grams of blood agar-based were mixed with 250 ml of distilled water. The agar was completely dissolved with heat and agitation and was autoclave for 15 min at 121°C. The agar was cooled down and 12.5 ml of defibrinated blood and gentamicin were added. After, the agar was poured into sterile Petri plates.
The nasal swabs were inoculated using the cotton swabs and further streaking was done using a sterilized metal inoculating loop. Alternatively, the supernatant of the prepared soil samples was inoculated into 5% sheep blood agar with gentamicin and ketoconazole using a sterile metal inoculating loop. Then, the plates were incubated at 37°C for 4 days and inspected daily.
The identity of Burkholderia sp. isolates were screened using bacterial morphology in gram-stained smears (bipolar staining or safety pin appearance) and colonial morphology (from smooth to wrinkled colonies).
Gram Staining
The suspected bacterial isolates were subjected to gram-staining for morphological identification. The smear of a purified bacterial isolate was prepared by emulsifying a colony in a drop of distilled water on a slide. Subsequently, the slide was heat-fixed. The slide was flooded with crystal violet for 60 sec and rinsed. Then, the slide was submerged with Gram’s iodine for 60 sec and rinsed. Then, the slide was decolorized with alcohol for not more than 10 sec. Following rinsing, the smear was stained with safranin for 30 sec. The slide was rinsed and blotted dry. The stained smear was examined under oil immersion at 1000 x magnification (Coico et al., 2009).
Molecular Identification
DNA was extracted from suspected bacteria from the nasal swab and soil samples. One loop-full of the bacterial colony (≈ 15 µL) was added in 500 µL of double-distilled water in a 1.5 mL microcentrifuge tube and vortex. The tubes were centrifuged for 1 min at 1000 rpm and were placed in a heat block at 95°C for 10 min. After, the samples were ready for PCR or stored at 4°C until further use (Zhang et al., 2005).
Extracted DNA samples were subjected to β-actin amplification through PCR using the protocol of Mingala et al. (2009). The samples were tested for the presence of the β-actin gene (housekeeping gene) to assure that genomic DNA was extracted from the samples (Figure 1).
The samples that were positive for the housekeeping gene were also subjected to PCR based on previously published primers targeting the 16s rDNA gene of the genus Burkholderia (Brett et al., 1997). The PCR mixture has a total volume of 12 µL containing 5.2 µL of sterile distilled water,
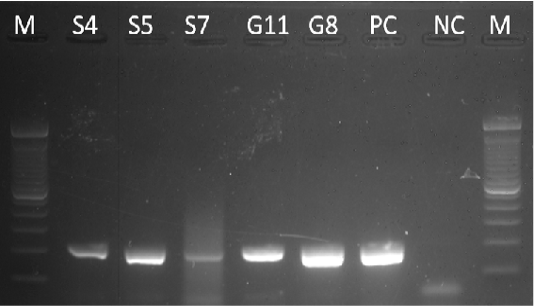
Figure 1: Gel analysis using the Beta-actin gene. M – 100 bp DNA ladder, PC – Positive Control, NC – Negative Control
2 µL of 5x PCR buffer (Promega, USA), 1.2 µL of 25 mM MgCl2 (Promega, USA), 0.5 µL of 2.5 mM dNTP (Intron, Korea), 0.1 µL of Taq Polymerase (Flexi Taq, Promega, USA), 0.5 µL of 10 µm each forward and reverses primers (Table 1) and 2 µL of DNA template. Thermal cycling conditions were initial denaturation of 95 °C for 1 min, followed by 25 cycles of denaturation of 95 °C for 15 sec, annealing of 62°C for 30 sec and extension of 72 °C for 30sec, and a final extension of 72 °C for 5 min. The PCR product was electrophoresed on a 2% agarose gel stained with GelRed (Biotium, USA). Positive samples have an amplicon size of 320 bp on the agarose gel.
Table 1: List of primers used in the study
| Primer | Primer Sequence | Reference |
| 16s rDNA | 5’ – AGACACGGCCCAGACTCCTAC – 3’ |
Brett et al. (1997) |
| 5’- CAGTCACCAATGCAGTTCCA – 3’ | ||
| Tat domain protein | 5’ – CAAGAACGGTTTATGCG -3’ |
Ho et al. (2011) Lau et al. (2014) |
| 5’- GAAGTGATCCATCAAATGTC – 3’ |
All 16s rDNA gene-positive samples were subjected to the amplification of the Tat-domain protein of Burkholderia sp. The PCR mixture has a total volume of 12 µL containing 5.2 µL of sterile distilled water, 2 µL of 5x PCR buffer (Promega, USA), 1.2 µL of 25 mM MgCl2 (Promega, USA), 0.5 µL of 2.5 mM dNTP (Intron, Korea), 0.1 µL of Taq Polymerase (Flexi Taq, Promega, USA), 0.5 µL of 10 µm each forward and reverse primers (Table 1) and 2 µL of DNA template. Thermal cycling was performed under the following condition: A hot start at 95 °C for 10 min, 10 touchdown cycles of 95 °C for 30 sec, annealing for 1.5 min at temperatures decreasing from 60 to 51 °C, 72 °C for 1 min, and 30 cycles of 95 °C for 30 sec, 50 °C for 1.5 min, 72°C for 1 min, and a final extension at 72 °C for 10 min (Lau et al., 2014; Ho et al., 2011). After PCR, products were subjected to gel electrophoresis using 2% agarose gel. The amplicon size of 189-bp is expected for the Tat-domain protein, which is specific for the pathogenic Burkholderia sp.
The products were resolved in 2% agarose gel stained in GelRed® (Biotium, USA). Three (3) µL of PCR products were loaded onto the wells in the agarose gel. Gel electrophoresis was undertaken for 30 min at an electrical current of 100 volts. The gel was then subjected to UV visualization using the Gel Documentation Machine (FlourChem E System, ProteinSimple, Inc., USA).
Tat-domain encoding gene PCR products were submitted to First Base, Malaysia for sequencing. Then, the sequences were assembled using MEGA 7 software. The aligned sequences were then subjected to the Basic Local Alignment Search Tool (BLAST) at the NCBI database to determine the sequence homology with the stored sequences. A phylogenetic tree was constructed using the neighbor-joining (NJ) method using MEGA 7 software. Confidence in the groups was estimated by a bootstrap of data using 1000 replications.
RESULTS AND DISCUSSION
Colony Morphology
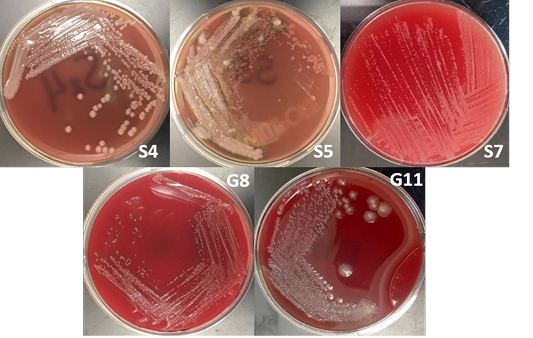
The bacterial isolates were subjected to colony morphological identification where shape, elevation, and surface characteristics were observed. There were 12 bacterial isolates for all samples: 8 from soil samples, 3 from goat nasal swab, and one from sheep nasal swab. However, there were only 5 isolates suspected as Burkholderia sp. based on cultural and morphological characteristics. These bacterial isolates are 3 and 2 bacterial isolates from soil and goat nasal swab samples, respectively (Figure 2).
Burkholderia sp. grows on most standard laboratory media, including blood agar. Burkholderia often reveals small, smooth, creamy colonies for the first 2 days and became wrinkled gradually after few days on blood agar (Gilligan and York, 2016; Cheng and Curie, 2005; Millan et al., 2007). The bacterium can be found in various sites including nasal and throat swabs, abscesses, and other biological components (Mohanty et al., 2016; Millan et al., 2007). Wuthiekanun (2015) reported that Burkholderia sp. normally presents resistance to gentamicin, presenting no zone and no latex-positive isolate with gentamicin susceptibility pattern. In addition, Burkholderia produces a distinct musty and earthy odor that is very pronounced on opening a Petri dish growing the microorganism or even opening an incubator door whenever a positive plate is present (Gilligan and York, 2016).
Morphological Characterization
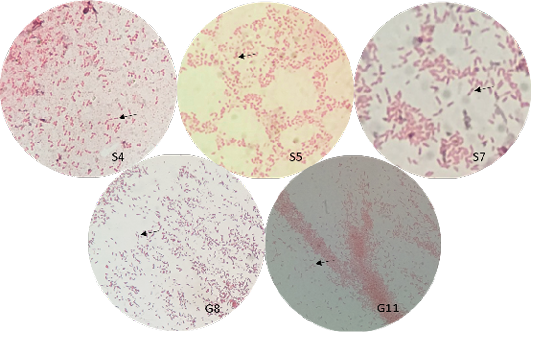
The bacterial isolates were observed under oil immersion at 1000x magnification, cell shape, and Gram-staining color of the bacteria was examined. The isolated bacteria from all samples (Figure 3) showed characteristic gram-negative, bacillus, and safety pin appearance (bipolar staining), which is one of the distinguishable characteristics of Burkholderia sp.
A non-fermenting bacterium, which was gentamicin-resistant and showed the typical “safety pin” appearance on gram-staining was previously identified that is under the Genus Burkholderia (Caldera et al., 2013; Garner et al., 2003; Putchery et al., 2009). Giligan et al. (2008) also describe the members of the genus as oxidase-positive, aerobic bacillus, and may be straight or slightly curved that is neither yellow nor violet pigmented. However, the bacterium may show different forms from older bacterial cultures and clinical samples (Garner et al., 2003). Furthermore, Burkholderia sp. can be differentiated from Pseudomonas aeruginosa through an oxidase test (Hemajarata et al., 2016) as Burkholderia sp. were former members of Pseudomonas spp.
Molecular Identification
The remaining 5 bacterial isolates undergone DNA extraction. These five extracted DNA was subjected to β-actin gene amplification (Figure 1). All positive extracted DNA samples for β-actin were used as the template for PCR amplification using 16S rDNA and the Tat-domain protein of the Genus Burkholderia.
Samples S4, S5, S7, G8, and G11 were subjected to PCR using the 16s rDNA gene. All samples exhibited an amplicon size of 320 bp (Figure 4). The sequence of a 320 bp portion of the 16s rDNA from pathogenic and nonpathogenic Burkholderia sp. compared to previously published sequences of Burkholderia sp. (Brett et al., 1997). The 16s rDNA gene is used for the study of the relatedness of prokaryotic species in terms of bacterial identification, species identification, and subtyping and identifying hypervirulent bacterial clones (Gee et al., 2003). In addition, the use of the 16s rDNA gene in the clinical laboratory is for ascertaining unidentified bacteria biochemically and providing reference identification for unusual strains (Patel, 2001). However, sequencing of 16s rDNA is unreliable and misidentifications still occur and are published (Janda and Abbott, 2007). This is also the reason for also targeting the Tat-domain protein of Burkholderia sp.
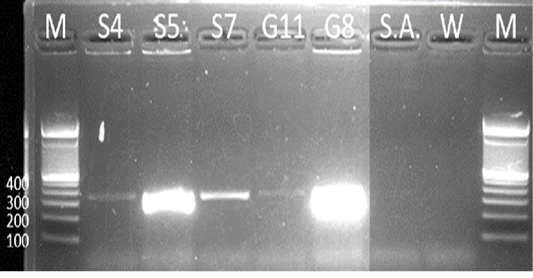
Figure 4: Gel analysis using Burkholderia-specific 16S rDNA. M - GeneRuler1kb plus DNA ladder. S.A – Staphylococcus aureus. W – Double Distilled Water
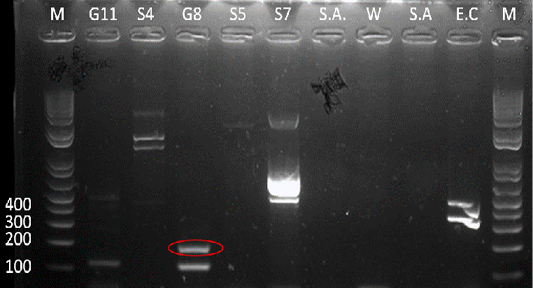
Figure 5: Gel analysis using Tat-domain encoding gene Burkholderia sp. primer. M - GeneRuler1kb plus DNA ladder. W – Double Distilled Water. S.A. – Staphyloccocus aureus. E.C. – Escherichia coli.
Table 2: Generated partial sequence of B. vietnamiensis strain G4 (sample G8) isolated in the Philippines
|
ID (Accession Number) |
Sequence | Amplicon size (bp) | Percent Identity |
|
G8 (MT452650) |
TTTATGCGGCGGTCTCGTT GGTTTCCCGGCAGTATTGAAACC GGTAGTTCGGTATCTGAATGTTGCT CAGTAAAATCAACGCGTTATGTGTTTTGTTCGATGCTTCG CAGCGGTAATTTTCGAACTCTGTCACTAATAGATGCTG CGATAATCGATGTTTTTTCGGCGTATCGACATTTGATG GATCACTTC |
192 |
97.86% (CP000614.1) |
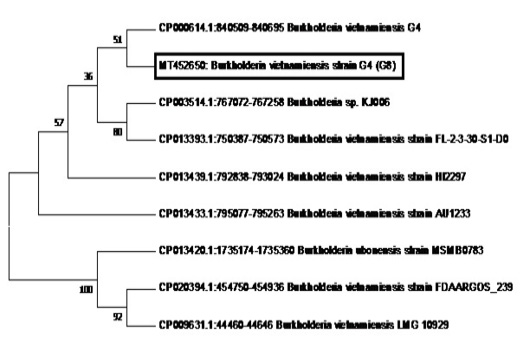
Figure 6: Phylogenetic tree showing the relationship of sample G8 (in the box with Accession Number MT452650) to B. vietnamiensis sequences registered in GenBank.
After the 16s rDNA gene amplification, all isolates were subjected to amplify the 189-bp fragment of the Tat-domain protein of Burkholderia sp. Only sample G8 exhibited an amplicon size of 189-bp (Figure 5), however, there are nonspecific bands to other samples and double band to sample G8. Thus, the purification of the 189-bp band was done and was sent for sequencing. Ho et al. (2011) performed a single-target PCR assay for detecting Burkholderia sp. targeting specifically the Tat-domain protein gene. In addition, a previous study proved through sequencing targeting the Tat-domain protein is specific for Burkholderia sp. (Lau et al., 2014). The Tat-domain protein was proved to be a sensitive and specific alternative for detecting Burkholderia sp. as compared to bacterial culture (Ho et al., 2011; Lau et al., 2014). Partial sequences of the sample G8 (Table 2) in this study showed 97.86% identity to Burkholderia vietnamiensis strain G4 (CP000614.1) registered in GenBank.
The phylogenetic analysis of the Tat-domain protein gene of sample G8 is shown in Figure 6 using the Neighbor-Joining method. Sample G8 formed a clade with B.vietnamiensis strain G4 (CP000614.1). Furthermore, most of the B. vietnamiensis isolates registered in GenBank showed 95% homology to sample G8.
B. vietnamiensis is part of the B. cepacia complex (BCC) that is responsible for a widespread and deadly respiratory disease like cystic fibrosis in immunocompromised individuals (Biddick et al., 2003; Hauser et al., 2011; Sawana et al., 2014).
With this finding, this is the first report of B. vietnamiensis was isolated in a nasal swab of a goat in the Philippines. B. cepacia complex genovar III and genovar V (B. vietnamiensis) was reported to have caused an outbreak of subclinical mastitis in a flock of dairy sheep. Since B. cepacia complex first association to subclinical mastitis, this will have important implications on one health as these bacteria are used as bacterial biopesticide and bioremediation (Berriatua et al., 2001).
The animals used in the study were apparently healthy and were maintained on the farm for more than a year. Furthermore, no new animals were introduced into the flock. Berriatua et al. (2001) reported that this may be due to environmental contamination.
Members of B. cepacia complex are difficult to culture and identify, thus, their impact on human and veterinary medicine may have been underappreciated and underestimated (Henry et al., 1996; Shelly et al., 2000). The study of Berriatua et al. (2001) reported the medical significance of the members of BCC in ovine mastitis. This may also be in the case of other ruminants such as goats, cattle, and water buffaloes. In addition, screening for Burkholderia sp. in animals with mastitis should be included in their screening test as this genus is resistant to most antibiotics (Rhodes and Scheweizer, 2016) and B. vietnamiensis is considered as an opportunistic pathogen that may cause cystic fibrosis in immunocompromised individuals (Drevinek and Mahenthiralingam, 2010; Jassem et al, 2011). Furthermore, a foodborne route of infection to humans may be possible due to previous reports of detection of BCC in raw milk (Uraz and Citak, 1998) and cheese (Smith et al., 1987). The bottom line is that there is still much medical and agricultural interest in the pathogenic potential of individual taxa and individual strains of BCC (Berriatua et al., 2001). Thus, further analysis of B. vietnamiensis and other possible isolates of the genus Burkholderia in the Philippines is important to fully understand the extent of their medical and agricultural importance and potential hazards.
CONCLUSION AND RECOMMENDATION
This research has successfully identified and isolated B. vietnamiensis in a goat nasal swab sample using microbiological assay using cultural, morphological, and molecular recognition of suspected bacterial isolates, and further validated by amplifying 16s rDNA and sequencing of the Tat-domain protein. In addition, this is the first report of B. vietnamiensis isolated in a nasal swab of goat in the Philippines. Furthermore, high-throughput sequence analysis is needed to further characterize B. vietnamiensis and Burkholderia isolates in the Philippines.
Conflict of interest
All authors declare not to have a conflict of interest.
ACKNOWLEDGMENTS
We thank the Philippine Carabao Center (PCC) for the support to finish the study. Deep appreciation is also extended to the DA Biotechnology Program for their support. Special thanks to all the staff of the Biosafety and Environment Section of PCC for their technical support.
authors contribution
Gabriel Alexis SP. Tubalinal and Danica C. Gregorio performed the research activity, compiled the data and prepared the manuscript. Gabriel Alexis SP. Tubalinal, Jerwin R. Undan and Claro N. Mingala designed the experiment, analyzed the data and prepared the manuscript. Claro N. Mingala for the final approval of the manuscript for publication. All authors have read and approved the manuscript before publication.
REFERENCES