Unlocking Insights into Saccharomyces cerevisiae and Milk Yields: A Meta-Analysis
Research Article
Unlocking Insights into Saccharomyces cerevisiae and Milk Yields: A Meta-Analysis
Bilal Ahmed1, Asep Setiaji1*, Lisa Praharani4, Faheem Ahmed Khan2, Nuruliarizki Shinta Pandupuspitasari3, Mohammad Miftakhus Sholikin4,5,6,7, Windu Negara2, Azhar Ali1, Muhammad Rizwan Yousaf1, Hamza Zulfiqar1, Alina Munawar8
1Animal Science Department, Diponegoro University, Semarang, Indonesia; 2Research Center for Animal Husbandry, National Research and Innovation Agency, Jakarta Pusat, 10340, Indonesia; 3Laboratory of Animal Nutrition and Feed Science, Animal Science Department, Faculty of Animal and Agricultural Sciences, Universitas Diponegoro, Semarang, Indonesia; 4Research Center for Animal Husbandry, Research Organization for Agriculture and Food, National Research and Innovation Agency (BRIN), Bogor, Indonesia; 5Meta-Analysis in Plant Science (MAPS) Research Group, Bandung 40621, Indonesia; 6Center for Tropical Animal Studies (CENTRAS), The Institute of Research and Community Empowerment of IPB (LPPM IPB), Bogor 16680, Indonesia; 7Animal Feed and Nutrition Modelling (AFENUE) Research Group, IPB University, Bogor 16680, Indonesia; 8Department of Biology, Government Associates College for Women China Scheme Lahore, Pakistan.
Abstract | Dairy farming’s sustainability thrives on profitable milk production, which demands an in-depth knowledge of influencing aspects. One of the dietary aspect involves Saccharomyces cerevisiae, a yeast that improves nutrient digestibility and milk yield. This meta-analysis, encompassing 44 research studies and 59 comparisons, investigates the link between Saccharomyces cerevisiae supplementation and milk yield in Holstein, Holstein-Friesian, and Holstein-Friesian x Thai Native cattle. Key factors such as milk yield, breed, and Saccharomyces cerevisiae dosage were retrieved and evaluated using OPENME and Minitab, with RSM and meta-analysis approaches. The results show that Saccharomyces cerevisiae has a statistically substantial and moderate effect on milk yield (Hedges’ d = 0.240). Sub-group analyses suggest breed-specific effects, and recommend optimal dosage of 173.1 g for increased milk output in dairy calves. The findings are useful for future research studies and farmers.
Keywords | Saccharomyces cerevisiae, Milk yield, Holstein, Feed supplementation
Received | February 01, 2024; Accepted | April 07, 2024; Published | June 25, 2024
*Correspondence | Asep Setiaji, Animal Science Department, Diponegoro University, Semarang, Indonesia; Email: asepsetiaji93@gmail.com
Citation | Ahmed B, Setiaji A, Praharani L, Khan FA, Pandupuspitasari NS, Sholikin MM, Negara W, Ali A, Yousaf MR, Zulfiqar H, Munawar A (2024). Unlocking insights into Saccharomyces cerevisiae and milk yields: A meta-analysis. Adv. Anim. Vet. Sci., 12(8):1517-1524.
DOI | https://dx.doi.org/10.17582/journal.aavs/2024/12.8.1517.1524
ISSN (Online) | 2307-8316
Copyright: 2024 by the authors. Licensee ResearchersLinks Ltd, England, UK.
This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
INTRODUCTION
Dairy farming is a major component of the global agricultural sector (Bojovic and McGregor, 2023), fulfilling the gap in the rising need for milk and dairy products (Milić et al., 2023). Sustaining adequate milk production in dairy cattle is critical to the industry’s future sustainability and financial sustainability (Bai et al., 2023). However, the multidimensional integrity of parameters regulating the milk production across different cattle breeds requires a careful assessment (Andrighetto et al., 2023).
An extensive amount of research has been dedicated to the fluctuations in the milk production in dairy cow (Wang et al., 2023), providing major insights into the impact of numerous variables. Previous research has examined concerns such as dietary management (Valldecabres et al., 2023; Orzuna-Orzuna et al., 2023) breeding approaches (Ziętara and Mirkowska, 2023; Jayawardana et al., 2023) and atmospheric conditions (Razzaghi et al., 2023; Toro-Ospina et al., 2023; Khan et al., 2023) establishing the foundation for comprehending the complexities of milk yield. Despite these advances, specifically in terms of feed supplementation, a full compilation of the existing research produces disparity in the findings, exposing a gap in our comprehension of how various factors interplay among dairy cattle.
Saccharomyces cerevisiae, is a key player in improving dairy cattle health (Garcia-Mazcorro et al., 2020), and meat quality (Williams et al., 2021; Dávila-Ramírez et al., 2020) and milk production (Imrich et al., 2021; Sun et al., 2021; Besseboua et al., 2017). Essentially, a live yeast strain, improves milk yield and quality while additionally improving nutrient digestibility (Phesatcha et al., 2020; Schlabitz et al., 2022). The essential mechanism is its ability to regulate the rumen environment and promote the abundance of ruminal cellulolytic bacteria (Anjum et al., 2018). This emphasizes the importance of Saccharomyces cerevisiae as a dietary supplement that improves cattle health and dairy production efficiency.
However, the findings of many of studies on Saccharomyces cerevisiae are fluctuating, some reporting massive positive effect while others reporting non-significant and even negative effect (Arambel and Kent, 1990; Zaworski et al., 2014). This meta-analysis attempts to address a gap in the literature by thoroughly reviewing and integrating data from a variety of studies reporting all sorts of positive, negative and even non-significant effects. The significance of this research arises from its ability to provide a comprehensive understanding of the Saccharomyces cerevisiae impacting milk yield in Holstein, Holstein-Friesian, and Holstein-Friesian x Thai Native cattle. Our meta-analysis intends to provide significant insights to dairy production by investigating potential sources of heterogeneity. The ultimate goal is to provide evidence-based methods for increasing milk yield and ensuring the continued success of dairy farming techniques.
MATERIALS AND METHODS
Eligibility criteria, search strategy, and data extraction
NCBI was used to conduct a literature search. During the search, the keywords ‘Saccharomyces cerevisiae’, ‘Holstein’, and ‘milk yield’ were employed. In addition, the following criteria were utilized to identify literature: (1) full-text publications published in English; (2) peer-reviewed published journals; (3) indexed conference proceedings; (4) direct comparison of control vs Saccharomyces cerevisiae diets; (5) Holstein cow study; and (6) milk yield and amount of Saccharomyces cerevisiae used in treatments. The preliminary searches yielded 100 potential references (Figure 2). Following the screening, 56 references were removed (Figure 1) since the studies did not meet the inclusion criteria. In the end, a total of 44 papers were utilized for data extraction and statistical analysis (Figure 3), with adherence to the PRISMA-P guidelines throughout the entire process (Moher et al., 2019; Page et al., 2021; Haddaway et al., 2022).
Data coding
The study coding included the primary characteristic milk yield per day in control groups, milk yield per day in treatment group, dairy cow breed, amount of Saccharomyces cerevisiae used in grams per day, number of control group animals, number of treatment group animals and the standard deviations. The mean value of main selected parameters (milk yield in control, milk yield in treatments and amount of Saccharomyces cerevisiae) were recorded and tabulated, and the units of measurement were homogenized for further data analysis.
Testing metadata and determining the optimal level
The primary idea of the study revolves around understanding the relationship between the daily milk production under influence of Saccharomyces cerevisiae through meta-analysis. The Response Surface Method was employed by using Minitab® v21.4.2 to determine the optimum amount of the yeast for maximal milk yield via a Contour Plot and Response Optimizer.
Meta-analysis of both overall and sub-group data
A standard meta-analysis was used to compare the effects of the control and Saccharomyces cerevisiae diets on milk yield. The assessment was carried out using the Hedges’ d effect size in OpenMEE software 2.0 (Wallace et al., 2017), which is capable of evaluating the impact of paired treatments and determining the effect size while accounting for changes in sample size, measurement units, and statistical test findings (Marín-Martínez and Sánchez-Meca, 2010). The Saccharomyces cerevisiae group was pooled to form the experimental group (E), whereas the control group was pooled to form (C). The effect size (d) was calculated using the following formula:
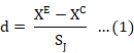
Where XC represents the mean value of control and XE denotes mean value of the experimental group. The measured parameter when in greater state, the effect size was positive, and vice versa. The SJ, is as follows, denotes small sample size correction factor:
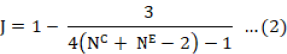
S shows the pooled standard deviation, formulated as:
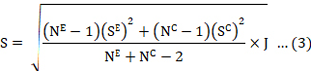
Where SE denotes the experimental group standard deviation and SC dor SD of control group, and NE denotes the experimental group’s sample size and NC denotes the control group’s sample size. The following is a description of Hedges’ d (Vd) variation:
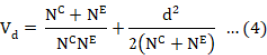
The cumulative effect size (d++) was computed as follows:
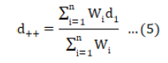
Where Wi is the sampling variance’s inverse: Wi=1⁄ Vd . The preciseness of the effect size, reported at 95% confidence interval (CI), represented by d++ ± (1.96 × SE), where SE (standard error) of cumulative effect size (J.S.M. and Marín-Martínez, 2011). If the computed effect size did not allign a null effect size, the result was statistically significant.
Publication bias
In order to uncover publication bias caused by non-significant papers that were excluded from the study, a fail-safe number (Nfs) was generated. A robust meta-analysis model was supposed to be Nfs (fail-safe number), which is greater than five times the study effect size used to compute the initial effect size (N) plus ten. Nfs was calculated using the Rosenthal approach (Rosenthal, 1979). The N Cohen benchmarks and the smallest sample size available from the relevant studies were utilized as reference points for effect size estimation. All of the aforementioned effect size computations were carried out with OpenMEE 2.0 (Wallace et al., 2017). In our analysis, the observed significance level was determined to be extremely tiny, particularly less than 0.0001, suggesting a highly significant impact. The Fail-Safe N is calculated to be 976, which means that approximately one thousand further insignificant trials would be needed in our analysis to potentially reduce the observed level of significance and get it nearer to the conventional significance criteria.
RESULTS and Discussion
Standard and cumulative meta-analysis
The standard and Cumulative Meta-analysis, employing the continuous random-effects model with DerSimonian-Laird as the random effects method, included data from 44 selected research studies that investigated the impact of a Saccharomyces cerevisiae intervention on variables such as Control Mean, Experimental Mean, Effect Size (Hedges’ d), and Variance. The cumulative results were examined step by step, adding studies sequentially and tracking the evolving effect size estimations. An effect size of 0.066, with later investigations changing the overall value. Notably, as further investigations were added, the cumulative impact size estimates developed more uniform. The overall effect size (Hedges’ d) was found to be 0.240, with a 95% confidence interval of (0.142, 0.339), demonstrating a statistically significant and moderate effect across the control and experimental groups in both standard and cumulative meta-analysis. The standard error was 0.050, and the p-value was below 0.001, underscoring the significance associated with the reported impact. Both of the meta-analysis revealed moderate heterogeneity across studies, with a Q-statistic of 96.723, a significant p-value (< 0.001), and an I² value of 41.069%. The calculated between-study variation (Tau-squared, τ²) was 0.038, indicating greater diversity in effect sizes than would be predicted by chance.
The forest plot (Figure 4) illustrates the outcomes of the standard meta-analysis and the forest plot (Figure 5) illustrates the outcomes of the cumulative meta-analysis which used Hedges’ d as the effect size measure to evaluate the impact of a given treatment on milk yield. Each horizontal line reflects an independent investigation, with squares representing point estimates and lines showing confidence intervals. The study weights, which are expressed as percentages, show the relative contribution of every research study to the total analysis. The overview beneath the plot indicates the cumulative effect, with the standardized mean difference, 0.240, with a 95% confidence interval of 0.142 to 0.339. This indicates the treatment’s significant positive impact on milk yield, as evidenced by the diamond dropping towards the right of the null line. The weights allocated to each trial in the cumulative analysis indicate how much they contribute to the total treatment effect, and the forest plot depicts rising consensus confirming the treatment’s effect on milk yield.
Meta-analysis of sub-group- breed
Th sub-group meta-analysis, employing Hedges’ d as the effect size measure, attempted to analyze the effects of particular treatments on milk yield while taking “breed” as a subgroup variable. The Continuous Random-Effects Model was adopted for this inquiry, with random effects addressed by the DerSimonian-Laird approach.
The subgroup analysis observed various results for various breeds as shown in forest plot (Figure 6). For Holstein cattle, the calculated effect size was 0.173, with a 95% confidence interval ranging 0.095 to 0.250, indicating an enormous positive influence on milk yield (p < 0.001). The effect size in the Holstein-Friesian subgroup was 0.392, but the confidence interval ranged from -0.261 to 1.045, showing that the treatment’s influence was less certain within this subgroup (p = 0.239). The Holstein-Friesian x Thai Native subgroup revealed a significant effect size of 4.013, with a confidence interval of 2.364 to 5.662 (p < 0.001), indicating a positive effect on milk yield across this breed.
The aggregate analysis, blending all subgroups, offered an estimated effect size of 0.240, with a 95% confidence interval spanning 0.142 to 0.339 (p<0.001). These findings point to the statistically significant and favorable overall effect of the therapy on milk yield throughout all breeds.
Control vs treatment means visual depiction
The alluvial diagram (Figure 7) depicts the fluctuating engagement of control and treatment milk yields among several observations. Each branching point denotes a unique set of paired values for control (C. Milk Yield) and treatment (T. Milk Yield). The fluctuating flow of data points demonstrates the range as well as distribution of the milk produced under both scenarios. There are clear transitions between control and treatment data, offering a visual narrative of the contrasting patterns in milk production. The (Figure 7) provides a brief but informative overview of the dataset, showing the variability in reactions to the experimental settings.
Optimal amount of Saccharomyces cerevisiae
The response surface optimization (Figure 8), which was performed using Minitab® v21.4.2, revealed that 173.152 grams of Saccharomyces cerevisiae is the ideal amount for maximum milk yield. The anticipated treatment milk yield for this ideal amount is 49.0262 (Table 1).
Table 1: RSM analysis using miniab for optimal dosage.
|
C. milk yield.x |
Amount (g) |
T. milk yield x Fit |
Composite desirability |
|
48 |
173.152 |
49.0262 |
1 |
This meta-analysis carried out to assess the effect of Saccharomyces cerevisiae supplementation on milk production in dairy cattle, thorough assessment of 44 research investigations revealed that Saccharomyces cerevisiae incorporates a significant influence on milk production. It supports and strengthens up prior research demonstrating the positive impacts on dairy production (Imrich et al., 2021; Sun et al., 2021; Besseboua et al., 2017). Saccharomyces cerevisiae supplementation improves milk output (Schlabitz et al., 2022), as indicated by an effect size of 0.240. This is backed up by Saccharomyces cerevisiae’s ability to enhance nutrient digestion and rumen health, resulting in increased milk production (Dávila-Ramírez et al., 2020). Subgroup analysis by breed offered further understanding on the differential effects of Saccharomyces cerevisiae supplementation. While the Holstein and Holstein-Friesian x Thai Native breeds had significant favorable effects on milk yield, the effect within the Holstein-Friesian subgroup was not as apparent. These breed-specific variations emphasize the need of taking into consideration genetic and physiological differences to assess nutritional interventions in future studies.
The current finding suggests ideal dosage of (173.152 grams per day) for maximum milk output and reinforces the meta-analysis’s practical relevance. This information can help dairy farmers optimize supplementing tactics to increase milk production more efficiently. Further research should explore additional factors influencing the effectiveness of Saccharomyces cerevisiae supplementation, such as environmental conditions, breed and management practices, to optimize milk production and enhance the sustainability of dairy farming systems. It is to be noted that the above findings are based on a specified parameters and current available studies, and further insights based on environmental conditions, lactation stages, age, etc. are the limitations and require future research efforts to fully open-up the way to comprehensive understanding of all associated factors.
CONCLUSIONs and Recommendations
In conclusion, this meta-analysis offers a thorough overview of the effect of Saccharomyces cerevisiae supplementation on milk output in Holstein, Holstein-Friesian, and Holstein-Friesian x Thai Native cattle. The study found a statistically significant and moderate total impact size, underscoring Saccharomyces cerevisiae’s positive influence on milk production. The conventional meta-analysis found that Saccharomyces cerevisiae had a consistent and positive influence. The cumulative meta-analysis, which tracked changing effect size estimates, verified the stability and significance of the determined treatment effect. The results showed a strong and consistent favorable influence on milk output across experiments. A subgroup meta-analysis by cattle breed found that Holstein cattle had a significant favorable impact on milk yield (effect size = 0.173, 95% CI [0.095, 0.250], p < 0.001). The Holstein-Friesian subgroup had a less certain affect (effect size = 0.392, 95% CI [-0.261, 1.045], p = 0.239), whereas the Holstein-Friesian x Thai Native subgroup had a strong beneficial effect (effect size = 4.013, 95% CI [2.364, 5.662], p < 0.001). The aggregate analysis of all subgroups confirmed Saccharomyces cerevisiae’s overall strong and positive impact on milk output. This meta-analysis adds vital insights to the dairy farming sector by consolidating previous research about Saccharomyces cerevisiae’s positive impact on milk production. The findings suggest using 173.1 g of Saccharomyces cerevisiae as a supplement in diet to increase milk yield in dairy cattle. Saccharomyces cerevisiae comes with environmental trade-off like increased methane emissions which should not be ignored, future research studies are recommended for influence of environment, genetics and management practices to fully understand the possible impact of these factors and ultimately open doors towards sustainable dairy practices.
Acknowledgement
The authors would like to thank reviewers who put their efforts to review this manuscript.
Novelty Statement
This meta-analysis examines at the effect of Saccharomyces cerevisiae supplementation on milk yield in distinct cattle breeds, making breed-specific recommendations for optimal dose and improving understanding of dietary components in dairy farming.
Author’s Contribution
BA, AS, LP, FAK, NSP, WN, and MMS conceptualized the meta-analysis. BA, FAK extracted the data. BA analyzed the data. BA and FAK wrote the paper. AS, LP, AA, MRY, HZ and AM helped revise this manuscript. All authors read and approved the final manuscript.
Conflict of interest
The authors have declared no conflict of interest.
References
Andrighetto I, Serva L, Fossaluzza D, Marchesini G (2023). Herd level yield gap analysis in a local scale dairy farming system: A practical approach to discriminate between nutritional and other constraining factors. Animals, 13(3). https://doi.org/10.3390/ani13030523
Anjum MI, Javaid S, Ansar MS, Ghaffar A (2018). Effects of yeast (Saccharomyces cerevisiae) supplementation on intake, digestibility, rumen fermentation and milk yield in Nili-Ravi buffaloes. Iran. J. Vet. Res., 19(2): 96–100.
Arambel MJ, Kent BA (1990). Effect of yeast culture on nutrient digestibility and milk yield response in early- to midlactation dairy cows. J. Dairy Sci., 73(6): 1560–1563. https://doi.org/10.3168/jds.S0022-0302(90)78825-X
Bai Z, Liu C, Wang H, Li C (2023). Evolution characteristics and influencing factors of global dairy trade. Sustain. (Switzerland), 15(2). https://doi.org/10.3390/su15020931
Besseboua O, Hama B, Jean-Luc HAA (2017). Review on the beneficial effects of omega-3 enriched eggs by dietary flaxseed oil supplementation. J. Istanbul Vet. Scı. Rev., 3(3): 89–94.
Bojovic M, McGregor A (2023). A review of megatrends in the global dairy sector: What are the socioecological implications? Agric. Hum. Values, 40(1): 373–394. https://doi.org/10.1007/s10460-022-10338-x
Dávila-Ramírez JL, Carvajal-Nolazco MR, López-Millanes MJ, González-Ríos H, Celaya-Michel H, Sosa-Castañeda J, Barrales-Heredia SM, Moreno-Salazar SF, Barrera-Silva MA (2020). Effect of yeast culture (Saccharomyces cerevisiae) supplementation on growth performance, blood metabolites, carcass traits, quality, and sensorial traits of meat from pigs under heat stress. Anim. Feed Sci. Technol., 267(February), 114573. https://doi.org/10.1016/j.anifeedsci.2020.114573
Garcia-Mazcorro JF, Ishaq SL, Rodriguez-Herrera MV, Garcia-Hernandez CA, Kawas JR, Nagaraja TG (2020). Review: Are there indigenous Saccharomyces in the digestive tract of livestock animal species? Implications for health, nutrition and productivity traits. Animal, 14(1): 22–30. https://doi.org/10.1017/S1751731119001599
Haddaway NR, Page MJ, Pritchard CC, McGuinness LA (2022). Campbell systematic reviews - 2022 - Haddaway - PRISMA2020 An R package and Shiny app for producing PRISMA 2020-compliant.pdf, pp. 1–14.
Imrich I, Čopík Š, Mlyneková E, Mlynek J, Haščík P, Kanka T (2021). The effect of Saccharomyces cerevisiae additive to cattle ration on milk yield of dairy cows. Acta Fytotech. Zoot., 24: 45–48. https://doi.org/10.15414/afz.2021.24.mi-prap.45-48
Jayawardana JMDR, Lopez-Villalobos N, McNaughton LR, Hickson RE (2023). Heritabilities and genetic and phenotypic correlations for milk production and fertility traits of spring-calved once-daily or twice-daily milking cows in New Zealand. J. Dairy Sci., 106(3): 1910–1924. https://doi.org/10.3168/jds.2022-22431
Khan FA, Pandupuspitasari NS, Huang C, Negara W, Ahmed B, Putri EM, Lestari P, Priyatno TP, Prima A, Restitrisnani V, Surachman M, Akhadiarto S, Darmawan IWA, Wahyuni DS, Herdis H (2023). Unlocking gut microbiota potential of dairy cows in varied environmental conditions using shotgun metagenomic approach. BMC Microbiol., 23(1): 1–11. https://doi.org/10.1186/s12866-023-03101-7
Marín-Martínez F, Sánchez-Meca J (2010). Weighting by inverse variance or by sample size in random-effects meta-analysis. Educ. Psychol. Measur., 70(1): 56–73. https://doi.org/10.1177/0013164409344534
Marín-Martínez JSM, (2011). Meta-analysis in psychological research.
Milić D, Novaković T, Tekić D, Matkovski B, Đokić D, Zekić S (2023). Economic sustainability of the milk and dairy supply chain: Evidence from Serbia. Sustainability, 15(21): 15234. https://doi.org/10.3390/su152115234
Moher D, Shamseer L, Clarke MGD (2019). Preferred reporting items for systematic review and meta-analysis protocols (prisma-p) 2015 statement. Jpn. Pharmacol. Therapeut., 47(8): 1177–1185.
Orzuna-Orzuna JF, Dorantes-Iturbide G, Lara-Bueno A, Chay-Canul AJ, Miranda-Romero LA, Mendoza-Martínez GD (2023). Meta-analysis of flavonoids use into beef and dairy cattle diet: Performance, antioxidant status, ruminal fermentation, meat quality, and milk composition. Front. Vet. Sci., 10. https://doi.org/10.3389/fvets.2023.1134925
Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, Shamseer L, Tetzlaff JM, Akl EA, Brennan SE, Chou R, Glanville J, Grimshaw JM, Hróbjartsson A, Lalu MM, Li T, Loder EW, Mayo-Wilson E, McDonald S, Moher D (2021). The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. Br. Med. J., pp. 372. https://doi.org/10.1016/j.ijsu.2021.105906
Phesatcha K, Phesatcha B, Wanapat M, Cherdthong A (2020). Roughage to concentrate ratio and Saccharomyces cerevisiae inclusion could modulate feed digestion and in vitro ruminal fermentation. Vet. Sci., 7(4): 1–13. https://doi.org/10.3390/vetsci7040151
Razzaghi A, Ghaffari MH, Rico DE (2023). The impact of environmental and nutritional stresses on milk fat synthesis in dairy cows. Domest. Anim. Endocrinol., 83. https://doi.org/10.1016/j.domaniend.2022.106784
Rosenthal R (1979). The file drawer problem and tolerance for null results. Psychol. Bull., 86(3): 638–641. https://doi.org/10.1037//0033-2909.86.3.638
Schlabitz C, Neutzling LD, Volken de Souza CF (2022). A review of Saccharomyces cerevisiae and the applications of its byproducts in dairy cattle feed: Trends in the use of residual brewer’s yeast. J. Cleaner Prod., 332 (June 2021). https://doi.org/10.1016/j.jclepro.2021.130059
Sun X, Wang Y, Wang E, Zhang S, Wang Q, Zhang Y, Wang Y, Cao Z, Yang H, Wang W, Li S (2021). Effects of Saccharomyces cerevisiae culture on ruminal fermentation, blood metabolism, and performance of high-yield dairy cows. Animals, 11(8). https://doi.org/10.3390/ani11082401
Toro-Ospina AM, Faria RA, Dominguez-Castaño P, Santana ML, Gonzalez LG, Espasandin AC, Silva JAIV (2023). Genotype–environment interaction for milk production of Gyr cattle in Brazil and Colombia. Genes Genom., 45(2): 135–143. https://doi.org/10.1007/s13258-022-01273-6
Valldecabres A, Branco-Lopes R, Bernal-Córdoba C, Silva-del-Río N (2023). Production and reproduction responses for dairy cattle supplemented with oral calcium bolus after calving: Systematic review and meta-analysis. JDS Commun., 4(1): 9–13. https://doi.org/10.3168/jdsc.2022-0235
Wallace BC, Lajeunesse MJ, Dietz G, Dahabreh IJ, Trikalinos TA, Schmid CH, Gurevitch J (2017). OpenMEE: Intuitive, open-source software for meta-analysis in ecology and evolutionary biology. Methods Ecol. Evol., 8(8): 941–947. https://doi.org/10.1111/2041-210X.12708
Wang A, Su G, Brito LF, Zhang H, Shi R, Liu D, Guo G, Wang Y (2023). Investigating the relationship between fluctuations in daily milk yield as resilience indicators and health traits in Holstein cattle. J. Dairy Sci., 107(3): 1535–1548. https://doi.org/10.3168/jds.2023-23495
Williams MS, Mandell IB, Bohrer BM, Wood KM (2021). The effects of feeding benzoic acid and/or live active yeast (Saccharomyces cerevisiae) on beef cattle performance, feeding behavior, and carcass characteristics. Trans. Anim. Sci., 5(4): 1–11. https://doi.org/10.1093/tas/txab143
Zaworski EM, Shriver-Munsch CM, Fadden NA, Sanchez WK, Yoon I, Bobe G (2014). Effects of feeding various dosages of Saccharomyces cerevisiae fermentation product in transition dairy cows. J. Dairy Sci., 97(5): 3081–3098. https://doi.org/10.3168/jds.2013-7692
Ziętara W, Mirkowska Z (2023). Concentration of dairy cow breeding and competitiveness of Polish farms specialized in milk production. Ann. Pol. Assoc. Agric. Agribus. Econ., XXV(2): 168–181. https://doi.org/10.5604/01.3001.0016.2867
To share on other social networks, click on any share button. What are these?






