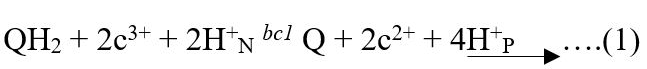Structural-Functional Characterization of Cytochrome b in bc1 and b6 f Complexes along with Polymorphic Analysis
Structural-Functional Characterization of Cytochrome b in bc1 and b6 f Complexes along with Polymorphic Analysis
Mohsin Shad1, Muhammad Usman2 and Qurratulann Afza Gardner1*
1School of Biological Sciences, University of the Punjab, Lahore, P.O. 54590, Pakistan.
2Department of Plant Pathology, University of Agriculture, Faisalabad, P.O. 38000, Pakistan
ABSTRACT
Mitochondrial cytochrome b (cyt b) transfers the electrons in bc1 complex from ubiquinone to cytochrome c reductase in the mitochondrial respiratory chain, and similarly in b6f enzyme complex, from plastoquinol (QH2) to plastocyanin (Pc) within the thylakoid membrane. Mitochondrial cyt b in bc1 complexes contains eight transmembrane helices A to H; two of these, B and E along with two histidines, are cross-linked with two hemes located at the top and bottom of the membrane. The b6f complex contains four large subunits which include cytochrome f, b, Rieske iron-sulfur protein, and subunit IV. Electrons transferred through these complexes are responsible for the pumping of protons across the membrane to produce ATP through ATP synthase. The present review provides the structural comparison of mitochondrial cyt b in ten model organisms targeting archaea, prokaryotes, and eukaryotes, highlighting phylogenetic and mutational analysis. Polymorphism in the mitochondrial cyt b gene helps in studying the biodiversity and is a valuable tool for the identification of species. Mutations in the cyt b gene produce abnormal protein leading to deficiencies in the complex III (coenzyme Q), resulting in defective oxidative phosphorylation and consequently effects other metabolic pathways. In the present article, we comprehensively compare the biodiversity of cyt b in bc1 and b6f complexes using in silico structural analysis tools, emphasizing that despite vast knowledge available in this field, still there are so much to explore about cyt b.
Article Information
Received 28 April, 2022
Revised 18 May 2022
Accepted 29 June 2022
Available online 20 September 2022
(early access)
Published 19 January 2023
Authors’ Contribution
MS has performed the literature review, data curation, and writing original draft. MU performed formal analysis and validation. QAG performed conceptualization, writing editing the final draft.
Key words
Mitochondria, Cytochrome b, bc1 complex, b6f complex, Mutational analysis, Conserved residues
DOI: https://dx.doi.org/10.17582/journal.pjz/20220428110433
* Corresponding author: qagardner.sbs@pu.edu.pk, annegardner80@yahoo.co.uk
0030-9923/2023/0002-975 $ 9.00/0
Copyright 2023 by the authors. Licensee Zoological Society of Pakistan.
This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
INTRODUCTION
Cytochrome b protein has a comprehensive role in the transportation of electrons in the respiratory chain of mitochondria. Cytochrome b is encoded by a single gene in the mitochondria, whereas in the case of chloroplast two genes are involved for the synthesis of this protein (Dumas et al., 2018). This polypeptide chain has a central role as electron transport in the bc1 and b6f transmembrane cytochrome complexes. Mitochondrial cytochrome b as a part of the bc1 complex transfers the electron from ubiquinone (QH2) to the cytochrome c reductase in the mitochondrial respiratory chain (Seddigh and Darabi, 2018). Four proton molecules (4H+P) are accumulated at the positive side of the lipid bilayer membrane by the oxidation of every ubiquinone (QH2) along with the reduction of two cyt c (2c3+) molecules (Equation 1). Mitochondrial cytochrome b present in the respiratory chain and chloroplast plastoquinol acceptor reductase (EC 7.1.1.6) are homologous in their function (Bhaduri et al., 2019). This chloroplast enzyme (b6f complexes) has a significant role in photosynthesis by transferring the electron from photosystem-II (PSII) to photosystem-I (PS-I). Moreover, this chloroplast enzyme contains two b and two c type cytochromes. The coenzyme Q also known as complex III is encoded by cytochrome b has a fundamental role in the production of ATP during oxidative phosphorylation (Stefely and Pagliarini, 2017).
Energy is required for the survival of all the organisms on this planet. All organisms possessing mitochondria except protozoa like Trychomonas contain cytochrome b and photo-redox proteins, which have a central role in ATP generation (Ramsay, 2019). ATP is renowned as a cellular currency for all living organisms provides energy to all the metabolic processes occurring in living cells. Some homologous proteins were also reported in the plant chloroplasts and cyanobacteria (cytochrome b6) that contribute their role in the b6f complex also known as plastoquinone–plastocyanin reductase complexes (EC 1.10.99.1) (Strand et al., 2017). The electrons transferred through these complexes are responsible for the pumping of protons across the membrane, produce ATP through membrane-bounded ATP synthase (Mansilla et al., 2018). ATP synthase works like a turbine, which is driven by the flow of protons from higher concentration to lower concentration across the membrane. ATP is generated from ADP by the addition of phosphate with ATP synthase which becomes active by proton motive force. ATP production mechanism in the electron transport chain located at the inner surface of the mitochondrial membrane is explained in Figure 1 (Nelson and Cox, 2017).
Structure of cytochrome b in mitochondrial bc1 complex
Cytochrome bc1 belongs to one of the three respiratory enzymes complexes located at the inner membrane in mitochondria. This complex transfers the electrons from ubiquinone to the cytochrome c reductase in the mitochondrial respiratory chain and uses energy from the electrochemical gradient across the membrane (Seddigh and Darabi, 2018). Cytochrome b in the mitochondrial genome is encoded by a single gene petB whereas in the chloroplast, encoded by petB and petD genes. The petD gene has a fundamental role in the production of subunit IV of the cyt b6f complex (ΔpetD) in the thylakoid membrane of the chloroplast (Dumas et al., 2018). The complete genome size of the mitochondrial cytochrome b gene is approximately 1140 bp (Lestari et al., 2018). The molecular mass of cytochrome b protein, along with hemes in the bc1 complexes, is approximately 42.5 kDa as reported by (Berry et al., 2000). The ATP production mechanism in the electron transport chain located at the inner surface of the mitochondrial membrane is explained in Figure 2 (Nelson and Cox, 2017). Cytochrome b from bc1 complex contains two ubiquinol/ubiquinone (Qo/Qi) and two hemes binding sites. Two hemes groups b562 and b566 are non-covalently attached to the cytochrome b in the mitochondria. Potentiometric titration showed that mitochondrial cytochrome b has two midpoint potential species included bH and bL. The species have higher midpoint potential represented by H while lower denoted by L. Two types of hemes bL and bH are present in the first helices bundles. Four histidine residues His84 and His183, His98 and His197 are present in the axial ligands of the bL and bH hemes and are highly conserved in the cytochrome b gene (Gao et al., 2003). The two species of hemes bH and bL have different properties and environments within the same protein.
The mitochondrial cytochrome b in bc1 complexes contains eight transmembrane helices A, B, C, D, E, F, G, and H (Dumas et al., 2018). Two transmembrane helices B and E in mitochondrial cyt b along with two histidine molecules are crosslinked with two hemes located at the top and bottom of the membrane. The 3D structure of mitochondrial cytochrome b in Mycobacterium tuberculosis is elaborated in Figure 3. The secondary structures of the mitochondrial cytochrome b are labelled with a spectrum ribbon along with N- and C-termini. The eight transmembrane helices of cyt b are labelled as A to H red alphabets (Fig. 3). The C and N termini of the mitochondrial cyt b are in the matrix region (Ko and Choi, 2016). The first five helices, A to E is surrounded by two hemes bH and bL subunits, are called first bundles. QN inhibitor site is present in the first bundle near to bH hemes site of the cyt b. The remaining three helices F, G, and H compose the second bundle of the cyt b. QP site is located near to the bL hemes is composed of helices C and F (Ko and Choi, 2016). The lipid bilayer of the mitochondrial membrane is represented as a doted red line (Fig. 3).
Structure of cytochrome b in chloroplast b6f complex
Cytochrome b6f complex exists as a dimer in the thylakoid membrane of the chloroplast. This complex contains four large subunits which included cytochrome f, cytochrome b, Rieske iron-sulfur protein, and subunit IV. Chloroplast b6f complex contains seven transmembrane helices in which the first four transmembrane helices A, B, C, and D are present in cytochrome b subunit of the b6f complex whereas subunit IV of the complex contains E, F, and G helices (Dumas et al., 2018). Different pet gene series is involved to produce the different subunits of the b6f complex. The petB gene is involved in the production of the cytochrome b6 of this complex whereas PetD is involved in subunit IV located at the thylakoid membrane of the chloroplast (Dumas et al., 2018). The cytochrome b6f oxidized the plastoquinol (PQH2) in the PSII just like the mitochondrial Q cycle. One electron is given to the bL hemes of cytochrome b6 while the other is passed to the Fe-S domain of the Rieske protein. The electrons from the PQH2 shifted to the plastocyanin which carries them to PSI (Tikhonov, 2018). The overall sketch of electron shifting and protons pumping mechanism was well elaborated in Figure 4 (Nelson and Cox, 2017).
Cytochrome b6 protein has two hemes binding domains represent as bL and bH. Two hemes groups b562 and b566 are non-covalently attached to the cytochrome b6 in the chloroplast thylakoid membrane. The total molecular weight of the cytochrome b6f complex is 217 kDa in which the cytochrome b6 subunit has 25 kDa. Approximately 215 amino acid is involved in the production of cytochrome b protein in the chloroplast. The molecular 3D structure of cytochrome b6f complex in alga Chlamydomonas reinhardtii is elaborated in Fig. 5 (MMDB ID# 25730) (https://www.ncbi.nlm.nih.gov/Structure/pdb/1Q90). The following b6f complex shows the structural similarities to bc1 complex of mitochondrial respiratory chain. This complex has some additional chlorophyll, beta-carotene, and haem binding sites.
Functions of cytochrome b in mitochondrial bc1 complex
The mitochondrial genes have a central role in the production of cytochrome b protein. This protein has a leading role in the synthesis of adenosine triphosphate (ATP). Mitochondrial cytochrome b protein converts the energy from food items into chemical energy. The electron transport chain located at the inner surface of the mitochondrial membrane is mainly involved in the synthesis of ATP (Letts and Sazanov, 2017). The respiratory chain located at the inner mitochondrial surface is composed of four different protein complexes, and free energy is generated by the pumping of protons across the membrane and by the transferring of electrons between the complexes (Manoj, 2018). This free energy is used for the generation of electrochemical gradients, across the mitochondrial membrane. The mitochondrial ATP synthase located at the inner mitochondrial surface generates ATP from ADP by the addition of phosphate. The proton motive force generated from electrochemical potential is used to run ATP synthase (Klusch et al., 2017).
The respiratory chain of the mitochondria possesses complexes I, II, III, and IV at the inner mitochondrial membrane. Complex III also known as bc1 complex, transfers the electrons from ubiquinone to the cytochrome c reductase in the mitochondrial respiratory chain (Zhu et al., 2020). Complex III is made up of membrane-bounded eleven subunits which included cytochrome b, cytochrome c, Rieske protein, two core proteins, and six low molecular weight proteins (Berry et al., 2000; Smith et al., 2004). The first three respiratory subunits are known as catalytic subunits. The remaining seven non-redox subunits are known as supernumerary subunits because they lack cofactor and are absent in the bacterial electron respiratory chain.
Complex III of the respiratory chain has a unique role in oxidative phosphorylation during which simple sugar is oxidized to produce ATP. The cytochrome b transfers the electrons particles to the complex III in the respiratory chain. Cytochrome b is the only component of the complex III that is encoded by the mitochondrial genome (Lestari et al., 2018). The mitochondrial cytochrome b gene region is extensively used in the phylogenetic relationship between different organisms due to its genetic variation. The genetic variation relationship within genera and families also determines through the cytochrome b gene segment. Comparative genomic results showed that mitochondrial DNA is rapidly evolved as compared to nuclear DNA. Therefore, the cytochrome b gene is a valuable marker for species identification and is being used as a marker in ticks, sand flies, mosquitoes, and tsetse flies.
Functions of cytochrome b in chloroplast b6f complex
Cytochrome b6f complex enzyme also known as plastoquinol-plastocyanin reductase (EC 1.10.99) is present at the thylakoid membrane of the chloroplast in many plants, green algae, and cyanobacteria (Bhaduri et al., 2019). This enzyme complex is responsible for the transferring of electrons from plastoquinol (QH2) to plastocyanin (Pc) within the thylakoid membrane (Kirchhoff et al., 2017). The reaction of electrons transferring in the b6f complex is functionally homologous to the bc1 complex reaction in the mitochondrial respiratory chain. The overall cytochrome b6f reaction mechanisms in cyclic and non-cyclic electrons transferring are explained below in the Equations 2 and 3.
H2O→PSII→QH2→Cyt b6f→Pc→PSI→NADPH (For non-cyclic pathway) …(2)
QH2→Cyt b6f→Pc→PSI→Q (For cyclic pathway) ….(3)
During photosynthesis, the reaction centres of photosystems I and II captures the light energy from the sun with the aid of multi subunits of cytochrome b6f complex. This complex transfers the electrons from PSII to PSI and pumps the protons across the membrane for the generation of Q-cycle. The reaction in the Q cycle in the chloroplast is like the complex-III of the mitochondrial respiratory chain. This free energy is used for the generation of electrochemical gradients across the thylakoid membrane in the chloroplast (Kanazawa et al., 2017). The ATP synthase located at the thylakoid membrane surface generates ATP from ADP by the addition of phosphate.
STRUCTURAL ANALYSIS OF CYTOCHROME B IN TEN MODEL ORGANISMS
Homology modelling and validation of cytochrome b in selected model organisms
The full-length protein sequences of mitochondrial cytochrome b gene, of ten model organisms, were retrieved from NCBI GenBank (https://www.ncbi.nlm.nih.gov) (Sayers et al., 2021). Ten model organisms were selected, ranging from archaea to mammals including Enterobacteriaceae BL21 (bacteria), Sulfolobus acidocaldarius N8 (archaea), Saccharomyces cerevisiae (yeast), Drosophila melanogaster (insect), Danio rerio (fish), Gallus gallus (bird), Homo sapiens (mammal; human), Mus musculus (mammal; rodent), Arabidopsis thaliana (plant), and Chlamydomonas reinhardtii (algae) are studied for structural and phylogenetic analysis. The 3D models of these selected cytochrome b were retrieved through online Alpha Fold Protein Structure Database developed by EMBL-EBI and DeepMind (https://alphafold.ebi.ac.uk/)(Fig. 6) (Jumper et al., 2021). The PDB file of selected cytochrome b models were opened and 3D models of the selected organisms were displayed into freely available Discovery Studio software (https://discover.3ds.com/discovery-studio-visualizer-download) (Studio, 2008). The model accuracy of cytochrome b in selected model organism were validated through online freely available SAVES server (https://saves.mbi.ucla.edu/). The overall quality factor for non-bonded atomic interaction of all these selected cytochrome b models of were determined through ERRAT server (https://servicesn.mbi.ucla.edu/ERRAT/) (Colovos and Yeates, 1993). The higher quality score of selected cytochrome b models indicating the higher quality of the models (Table I).
The position of alpha helices in membrane protein is determined through observing the Phyre2 generated models (Fig. 6). The locations of N- and C- termini were also observed through a schematic diagram generated through this Phyre 2 web tool (Fig. 6).
The stereochemical properties of overall structural geometry of selected cytochrome b models were predicted through PROCHECK server (https://servicesn.mbi.ucla.edu/PROCHECK/) (Laskowski et al., 1993). The Ramachandran plot check the stereochemical quality of the protein structure by analysing the geometry between the torsional angles ψ and φ of the residues within the peptide. The stereochemical properties of selected cytochrome b models are good when more than 90% of amino acid residues are in the most favour able region of the Ramachandran plot (Fig. 7). The overall quality factor, amino acid residues percentages in most favoured region, additional allowed region, generally allowed region and disallowed region of Ramachandran plot are shown in Table I. The secondary structures of cytochrome b in all selected model organisms were predicted through online Protein Homology/analogy Recognition Engine (Phyre 2) tool (http://www.sbg.bio.ic.ac.uk/servers/phyre2/html/page.cgi?Id=index) (Kelley et al., 2015). The position of alpha helices in membrane protein is determined through observing the Phyre2 generated models (Fig. 6). The locations of N- and C- termini were also observed through a schematic diagram generated through this Phyre 2 web tool (Fig. 6).
Phylogenetic analysis of cytochrome b in selected model organisms
To check the conserved structural residues, the protein sequences of mitochondrial cytochrome b membrane protein are aligned through the online available Clustal Omega tool (https://www.ebi.ac.uk/Tools/msa/clustalo) (Sievers et al., 2011). The position of alpha-helix residues in the aligned protein sequences is highlighted in yellow colour (Fig. 8). Maximum (A to K) transmembrane alpha-helix is observed in archaeal strain Sulfolobus acidocaldarius N8 and a minimum of (E, F, G, I) transmembrane helices are observed in prokaryotic Enterobacteriaceae BL21 strain with less homology with other selected model organisms. Homo sapiens and Drosophila melanogaster have (A to I) transmembrane helices in the mitochondrial cytochrome b transmembrane protein. The remaining six model organisms; yeast, starfish, bird, mammals, plants, and algae contain (A to I) transmembrane alpha-helices except helix D in the mitochondrial cytochrome b protein (Table II). The first transmembrane alpha-helix has glycine, leucine, and threonine residues (G, L, T) highlighted in turquoise colour remained conserved in archaea, unicellular, and multicellular eukaryotes organisms (Table II), whereas the conserved amino acids like tryptophan, leucine, glutamine, and alanine (W, L, Q, A) highlighted in gray shade are only conserved in eukaryotic organisms (Table III).
Table I. Validation of cytochrome b in selected model organisms through ERRAT, and PROCHECK.
|
Model organisms |
Errat |
Procheck |
|||
|
Quaility factor |
Most favoured region |
Additional allowed region |
Generally allowed region |
Disallowed region |
|
|
Enterobacteriaceae BL21 |
97.19% |
96.20% |
3.10% |
0.00% |
0.60% |
|
Sulfolobus acidocaldarius N8 |
88.87% |
84.70% |
12.70% |
1.70% |
0.90% |
|
Saccharomyces cerevisiae |
98.93% |
94.70% |
4.70% |
0.00% |
0.60% |
|
Drosophila melanogaster |
95.94% |
94.50% |
5.20% |
0.00% |
0.30% |
|
Danio rerio |
97.04% |
94.20% |
5.50% |
0.00% |
0.30% |
|
Gallus gallus |
92.45% |
85.70% |
13.40% |
0.60% |
0.30% |
|
Homo sapiens |
96.50% |
94.60% |
5.10% |
0.00% |
0.30% |
|
Mus musculus |
96.24% |
94.00% |
5.70% |
0.00% |
0.30% |
|
Arabidopsis thaliana |
94.50% |
92.90% |
6.80% |
0.00% |
0.30% |
|
Chlamydomonas reinhardtii |
84.98% |
93.80% |
5.50% |
0.00% |
0.60% |
Table II. Domain and transmembrane helices of mitochondrial cytochrome b protein in the model organism. Humans, insects, and Archaea have some additional transmembrane alpha-helices as compared to other model organisms. All the model’s organisms have N and C- terminal domains in the cytoplasm except humans, insects and archaea have extracellular N-terminal domains.
|
S. No |
Cytochrome B in different model organisms |
Classification |
No. of helices |
Domains |
|
|
N-terminal |
C-terminal |
||||
|
1 |
Enterobacteriaceae BL21 |
Bacteria |
4 |
Cytoplasmic |
Cytoplasmic |
|
2 |
Sulfolobus acidocaldarius N8 |
Archaea |
11 |
Extracellular |
Cytoplasmic |
|
3 |
Saccharomyces cerevisiae |
Yeast |
8 |
Cytoplasmic |
Cytoplasmic |
|
4 |
Drosophila melanogaster |
Insect |
9 |
Extracellular |
Cytoplasmic |
|
5 |
Danio rerio |
Starfish |
8 |
Cytoplasmic |
Cytoplasmic |
|
6 |
Gallus gallus |
Bird |
8 |
Cytoplasmic |
Cytoplasmic |
|
7 |
Homo sapiens |
Human |
9 |
Extracellular |
Cytoplasmic |
|
8 |
Mus musculus |
Mammal |
8 |
Cytoplasmic |
Cytoplasmic |
|
9 |
Arabidopsis thaliana |
Plant |
8 |
Cytoplasmic |
Cytoplasmic |
|
10 |
Chlamydomonas reinhardtii |
Algae |
8 |
Cytoplasmic |
Cytoplasmic |
The Enterobacteriaceae species lack helices from one to four (A to D) in multiple sequence alignment results. Structural analysis of the cyt b in model organisms from various kingdoms show that conserved amino acids are mostly located near N- and C-termini residues of the transmembrane alpha-helices. Some residues are also conserved within the helices for e.g., glycine, alanine, and histidine (G, A, H) are conserved within the second transmembrane helices in archaea and eukaryotes while serine, phenylalanine, glycine, and tyrosine (S, F, G, Y) are conserved only in the unicellular and multicellular model organism. The conserved residues in all the helices are highlighted in the multiple sequence alignment results. One interesting feature is that helix D is only observed in humans, archaea, and drosophila which might be the role in coping with extreme environmental conditions. The conserved residues asparagine, leucine, and proline (N, L, P) in helix D are highlighted with dark yellow colour. The conserved residues have a comprehensive role in membrane stability and evolution. The high number of transmembrane helices have a significant role in membrane stabilization. Archaea has two additional helices (J and K) as compared to humans or other eukaryotic organisms which might be leading factors to survive within extreme environmental conditions (Table III; Sr number: (2) The phylogenetic tree of mitochondrial cytochrome b, in all these model organisms, is generated through freely available MEGA-7 software (Fig. 9).
Table III. Conserved residues in the transmembrane alpha-helices of mitochondrial cytochrome b protein in the model organism.
|
Sr. No |
Cytochrome b in model organisms |
Conserved residues in transmembrane alpha-helices of mitochondrial cytochrome b |
||||||||||
|
A |
B |
C |
D |
E |
F |
G |
H |
I |
J |
K |
||
|
Enterobacteriaceae BL21 |
Absent |
Absent |
Absent |
Absent |
H L |
No match |
No match |
Absent |
No match |
Absent |
Absent |
|
|
2 |
Sulfolobusa acidocaldarius N8 |
G L T W L Q A |
G A H S F G Y |
W G A Y T F |
N L P |
H F H L |
K D P |
P G V |
L G |
G Q F |
Present |
Present |
|
3 |
Saccharomyces cerevisiae |
G L T W L Q A |
G A H S F G Y |
W G A Y T F |
ABSENT |
H F H L |
K D P |
P G V |
L G |
G Q F |
Absent |
Absent |
|
4 |
Drosophila melanogaster |
G L T W L Q A |
G A H S F G Y |
W G A Y T F |
N L P |
H F H L |
K D P |
P G V |
L G |
G Q F |
Absent |
Absent |
|
5 |
Danio rerio |
G L T W L Q A |
G A H S F G Y |
W G A Y T F |
ABSENT |
H F H L |
K D P |
P G V |
L G |
G Q F |
Absent |
Absent |
|
6 |
Gallus gallus |
G L T W L Q A |
G A H S F G Y |
W G A Y T F |
ABSENT |
H F H L |
K D P |
P G V |
L G |
G Q F |
Absent |
Absent |
|
7 |
Homo sapiens |
G L T W L Q A |
G A H S F G Y |
W G A Y T F |
N L P |
H F H L |
K D P |
P G V |
L G |
G Q F |
Absent |
Absent |
|
8 |
Mus musculus |
G L T W L Q A |
G A H S F G Y |
W G A Y T F |
ABSENT |
H F H L |
K D P |
P G V |
L G |
G Q F |
Absent |
Absent |
|
9 |
Arabidopsis thaliana |
G L T W L Q A |
G A H S F G Y |
W G A Y T F |
ABSENT |
H F H L |
K D P |
P G V |
L G |
G Q F |
Absent |
Absent |
|
10 |
Chlamydomonas reinhardtii |
G L T W L Q A |
G A H S F G Y |
W G A Y T F |
ABSENT |
H F H |
K D P |
P G V |
L G |
G Q F |
Absent |
Absent |
The conserved residues in archaea and eukaryotic are highlighted with turquoise colour. The eukaryotic conserved residues are highlighted with gray colour. The dark yellow colour indicates the conserved residues in the high number of transmembrane alpha-helices model organisms.
MITOCHONDRIAL CYTOCHROME B BASED PHYLOGENETIC AND MUTATIONAL ANALYSIS
Polymorphic effects of cytochrome b gene on fertility
The mutation in mitochondrial cytochrome b gene has significant effects on the reproductive system of an organism. Genetic diseases are mostly associated with the substitution and deletions of bases in the DNA (Pal et al., 2019). The mitochondrial genome is inherited matrilineally while the nuclear genome is inherited from both mother and father (Wolf et al., 2017). Energy is required for the proper functioning of reproductive system of living organisms (Haas et al., 2019). The mutation in the cytochrome b gene leads to disturbance in oxidative phosphorylation in mitochondria and consequently other metabolic pathways. In the male reproductive system, the propelling and moving processes of sperm utilizes energy from mitochondria, located at the base of the flagellum (Cardullo and Baltz, 1991). The reduction of energy metabolism leads to defects in male fertility. In Drosophila melanogaster the male infertility is observed due to single amino acid substitution from alanine to threonine at position 278 in the protein expressed by cytochrome b gene (Patel et al., 2016). This mutation was also observed in the cytochrome b of many other species of vertebrates and invertebrates. The variation in the mitochondrial encoded gene is also responsible for the shorter lifespan of the males in many taxa (Patel et al., 2016).
The polymorphism in mitochondrial cytochrome b gene is extensively used for phylogenetic and biodiversity analysis (Jadav et al., 2013; Mai et al., 2014; Saikia et al., 2015). The mutation in the cytochrome b gene affects the male and female reproductive system because it has an equal contribution in transferring and providing the energy in the development of zygote, embryo, and oocytes in females (Ramalho-Santos et al., 2009). The mutation in the cytochrome b gene is responsible for the alternation in the post-translation modification like the occurrence of transmembrane helices, phosphorylation site, and leucine-rich nuclear export signal (Pradhan et al., 2018). Recent research has shown that the mutation in the cytochrome b gene induces malfunctioning of the female reproduction of Ghungroo pig. The application of this study has been used as a valuable marker for studying the Ghungroo pig female reproductive system (Pradhan et al., 2018).
Cytochrome b gene role in species determination
The mitochondrial cytochrome b gene has a tremendous role in species determination throughout the world. The trading business of endangered species across the world is confirmed through the amplification of the cytochrome b gene. The mitochondrial cytochrome b gene has been extensively sequenced among all the vertebrates. Cytochrome b gene is famous in the scientific world due to its use as a tracer in the phylogenetic relationship (Cutarelli et al., 2018). Species identification through mitochondrial cytochrome b gene has a wonderful role in criminal problems, paternity, and maternity testing, illegal trade, lineage, and inbreed counting (Reid, 2018). In many parts of the world species identification is done through the cytochrome b gene (Cutarelli et al., 2018). The mitochondrial genome has comparatively less mutational change as compared to the nuclear genome and remains conserved throughout the species. Different mitochondrial genes are involved in species identification like 12S, 16S rRNA, and cytochrome b gene. The cytochrome b gene is extensively used for species identification due to the presence of the stable and variable sequences used by the universal primers. The complete genome size of the mitochondrial cytochrome b gene is approximately 1140 bp (Rahat et al., 2020). The amplification of the whole gene is difficult, so the short fragments like 402 bp and 359 bp of the cytochrome b gene is used. These fragments are generated through AluI restriction endonuclease. Different restriction endonucleases have been reported that are used in the cleavages of cytochrome b gene like BsaJI, AluI, BstUI, TaqI, and BstNI, HinfI, RsaI, and NsiI. The highlighting feature of the cytochrome b gene possesses high polymorphism which makes them key player genes in species identification.
Health conditions associated with cytochrome b gene
The mitochondrial genes are responsible for the production of cytochrome b protein of mitochondrial bc1complex.The mutations in the complex III encoding genes are responsible for the deficiencies of respiratory enzymes in mitochondria. The deficiencies of this complex III subunit of the mitochondrial respiratory chain lead to neuromuscular and movement disorders in human beings (Bénit et al., 2009). The mitochondrial complex III is inherited from the nuclear and matrilineal mode of origin. The complex III respiratory enzyme is produced from one of the mitochondrial cytochrome b gene and nine nuclear genes BCS1L, UQCRQ, TTC19, UQCRB, UQCRC2, CYC1, LYRM7, UQCC2, and UQCC3 (Wanschers et al., 2014). Five genes BCS1L, LYRM7, TTC19, UQCC2, and UQCC3 are assembly factors that are involved in the assembly of the complex III subunit of the respiratory chain (Invernizzi et al., 2013; Tucker et al., 2013). The main highlighting feature of this mutational complex III encoding enzymes is the failure to thrive, bilateral retinal cherry-red spots, and progressive neurodegeneration with Leigh-like brain MRI abnormalities (Mordaunt et al., 2015). All the mutations linked to the TTC19 gene are nonsilent and have a significant contribution to the deficiencies of mitochondrial complex III respiratory enzyme. The severity of the complex III deficiency is linked to the mutation in the mitochondrial cytochrome b gene (Mordaunt et al., 2015). The mutated mitochondrial DNA is present with a high percentage in the skeletal muscles which is the clue to identify the myopathies in this individual.
CONCLUSION
In this review article, structural and functional analysis of cytochrome b and their significant contribution in mitochondrial respiratory chain bc1 and thylakoid membrane of the chloroplast containing b6f complexes are studied. The present review highlights that the different pet gene is involved in the synthesis of cytochrome b protein. The mitochondrial cytochrome b is encoded by a single petB gene whereas in chloroplast petB and petD gene is involved in cytochrome b production. The petD gene has a fundamental role in the production of subunit IV of the cyt b6f complex (ΔpetD) in the thylakoid membrane of the chloroplast. The mitochondrial cytochrome b in bc1 complexes contains eight transmembrane helices A, B, C, D, E, F, G, and H. Two transmembrane helices B and E in mitochondrial cyt b along with two histidine molecules are crosslinked with two hemes located at the top and bottom of the membrane. Chloroplast b6f complex contains seven transmembrane helices in which the first four transmembrane helices A, B, C, and D are present in cytochrome b subunit of the b6f complex whereas subunit IV of the complex contains E, F, and G helices. The reaction of electrons transferring in the b6f complex is functionally homologous to the bc1 complex reaction in the mitochondrial respiratory chain. The electrons transferred through these complexes are responsible for the pumping of protons across the membrane-produced ATP through membrane-bounded ATP synthase.
The present review highlights the in silico structure analysis of mitochondrial cytochrome b protein from ten different model organisms is studied starting from bacteria, archaea, yeast, bird, fish, insect, algae, mammal, plant, and humans are predicted through the Alpha Fold 2 and phyre-2 web tool. A detailed structural comparison and sequence alignment of mitochondrial cytochrome b membrane protein is studied through a web-based bioinformatics tool in Enterobacteriaceae BL21, Sulfolobus acidocaldarius N8, Saccharomyces cerevisiae, Drosophila melanogaster, Danio rerio, Gallus gallus, Homo sapiens, Mus musculus, Arabidopsis thaliana, and Chlamydomonas reinhardtii model organisms. The conserved residues in the transmembrane alpha-helices are predicted through multiple sequence alignment tools. The conserved residues have a comprehensive role in membrane stability and evolution. The high number of transmembrane helices has a significant role in membrane stabilization. Archaea have two additional helices as compared to humans or other eukaryotic organisms which might be leading factors to survive within extreme environmental conditions.
The mitochondrial cytochrome b gene is used as a valuable tool in species identification and phylogenetic analysis throughout the world. The mitochondrial genome has comparatively less mutational change as compared to the nuclear genome and remains conserved throughout the species. The cytochrome b gene is extensively used for species identification due to the presence of the stable and variable sequences used by the universal primers. The mutation in the cytochrome b gene produces a short and abnormal protein which leads to the deficiencies of the complex III protein has some impact reproductive system. The deficiencies of this complex III subunit of the mitochondrial respiratory chain leads to neuromuscular and movement disorders in human beings. In silico structure, analysis helps the scientist to better understand the advancement in the evolutionary process. The inheritance of the individuals is strongly linked with the mitochondrial genome. The genome editing through CRISPR/CAS9 technology is the state-of-the-art technology to remove the mutated DNA sequences within the genome. Using this technique, the harmful mutations in mitochondrial cyt b can be edited which opens a new horizon of research to unravel this field.
ACKNOWLEDGEMENTS
We are highly thankful to Prof. Dr Muhammad Akhtar (FRS) for his helpful comments on the manuscript.
Funding
This research received no external funding.
Informed consent statement
The information utilized in this review paper is public access and was appropriately cited for recognition of the original authors. Specific informed consent was not deemed necessary
Statement of conflict of interest
The authors have declared no conflict of interest.
REFERENCES
Bénit, P., Lebon, S. and Rustin, P., 2009. Respiratory-chain diseases related to complex iii deficiency. Biochim. Biophys. Acta Mol. Cell Res., 1793: 181-185. https://doi.org/10.1016/j.bbamcr.2008.06.004
Berry, E.A., Guergova-Kuras, M., Huang, L.S., and Crofts, A.R., 2000. Structure and function of cytochrome bc complexes. Annu. Rev. Biochem., 69: 1005-1075. https://doi.org/10.1146/annurev.biochem.69.1.1005
Bhaduri, S., Zhang, H., Erramilli, S. and Cramer, W.A., 2019. Structural and functional contributions of lipids to the stability and activity of the photosynthetic cytochrome b6f lipoprotein complex. J. biol. Chem., 294: 17758-17767. https://doi.org/10.1074/jbc.RA119.009331
Cardullo, R.A. and Baltz, J.M., 1991. Metabolic regulation in mammalian sperm: Mitochondrial volume determines sperm length and flagellar beat frequency. Cell Motil. Cytoskeleton, 19: 180-188. https://doi.org/10.1002/cm.970190306
Colovos, C. and Yeates, T.O., 1993. Verification of protein structures: Patterns of nonbonded atomic interactions. Protein Sci., 2: 1511-1519. https://doi.org/10.1002/pro.5560020916
Cutarelli, A., Galiero, G., Capuano, F., and Corrado, F., 2018. Species identification by means of mitochondrial cytochrome b DNA sequencing in processed anchovy, sardine and tuna products. Fd. Nutr. Sci., 9: 369. https://doi.org/10.4236/fns.2018.94029
Dumas, L., Zito, F., Auroy, P., Johnson, X., Peltier, G. and Alric, J., 2018. Structure-function analysis of chloroplast proteins via random mutagenesis using error-prone pcr. Pl. Physiol., 177: 465-475. https://doi.org/10.1104/pp.17.01618
Gao, X., Wen, X., Esser, L., Quinn, B., Yu, L., Yu, C.A., and Xia, D., 2003. Structural basis for the quinone reduction in the bc 1 complex: A comparative analysis of crystal structures of mitochondrial cytochrome bc 1 with bound substrate and inhibitors at the qi site. Biochemistry, 42: 9067-9080. https://doi.org/10.1021/bi0341814
Haas, J., Bassil, R., and Casper, R.F., 2019. The role of mitochondria in reproductive function and assisted reproduction. In: The ovary. Elsevier, pp. 337-342. https://doi.org/10.1016/B978-0-12-813209-8.00021-2
Invernizzi, F., Tigano, M., Dallabona, C., Donnini, C., Ferrero, I., Cremonte, M., Ghezzi, D., Lamperti, C., and Zeviani, M., 2013. A homozygous mutation in lyrm 7/mzm 1 l associated with early onset encephalopathy, lactic acidosis, and severe reduction of mitochondrial complex iii activity. Hum. Mutat., 34: 1619-1622. https://doi.org/10.1002/humu.22441
Jadav, K.K., Shrivastav, A.B., and Rajput, N., 2013. Development of molecular tools to differentiate indian wild pig (Sus scrofa cristatus) meat from exotic and local domestic pig meat. Vet. World, 6: 919. https://doi.org/10.14202/vetworld.2013.919-922
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., Tunyasuvunakool, K., Bates, R., Žídek, A., and Potapenko, A., 2021. Highly accurate protein structure prediction with alphafold. Nature, 596: 583-589. https://doi.org/10.1038/s41586-021-03819-2
Kanazawa, A., Ostendorf, E., Kohzuma, K., Hoh, D., Strand, D.D., Sato-Cruz, M., Savage, L., Cruz, J.A., Fisher, N., and Froehlich, J.E., 2017. Chloroplast atp synthase modulation of the thylakoid proton motive force: Implications for photosystem i and photosystem ii photoprotection. Front. Pl. Sci., 8: 719. https://doi.org/10.3389/fpls.2017.00719
Kelley, L.A., Mezulis, S., Yates, C.M., Wass, M.N., and Sternberg, M.J., 2015. The phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc., 10: 845-858. https://doi.org/10.1038/nprot.2015.053
Kirchhoff, H., Li, M., and Puthiyaveetil, S., 2017. Sublocalization of cytochrome b6f complexes in photosynthetic membranes. Trends Pl. Sci., 22: 574-582. https://doi.org/10.1016/j.tplants.2017.04.004
Klusch, N., Murphy, B.J., Mills, D.J., Yildiz, Ö., and Kühlbrandt, W., 2017. Structural basis of proton translocation and force generation in mitochondrial atp synthase. Elife, 6: e33274. https://doi.org/10.7554/eLife.33274.001
Ko, Y., and Choi, I., 2016. Putative 3d structure of qcrb from Mycobacterium tuberculosis cytochrome bc 1 complex, a novel drug-target for new series of antituberculosis agent q203. Bull. Korean chem. Soc., 37: 725-731. https://doi.org/10.1002/bkcs.10765
Laskowski, R.A., MacArthur, M.W., Moss, D.S., and Thornton, J.M., 1993. Procheck: A program to check the stereochemical quality of protein structures. J. appl. Crystallogr., 26: 283-291. https://doi.org/10.1107/S0021889892009944
Lestari, D.A., Purbowati, E., Sutopo, S., and Kurnianto, E., 2018. Amino acid sequence based on cytochrome b gene in kejobong goat and its genetic relationships among several local goats in asia. Vet. World, 11: 1196. https://doi.org/10.14202/vetworld.2018.1196-1202
Letts, J.A., and Sazanov, L.A., 2017. Clarifying the supercomplex: The higher-order organization of the mitochondrial electron transport chain. Nat. Struct. Mol. Biol., 24: 800-808. https://doi.org/10.1038/nsmb.3460
Mai, N.T.P., Chung, D.C., Chi, H.N.Q. and Son, H.N., 2014. The genetic relationship of vietnamese pigs in central highlands assessed by cytochrome b. Open J. Genet., 2014. https://doi.org/10.4236/ojgen.2014.45033
Manoj, K.M., 2018. Aerobic respiration: Criticism of the proton-centric explanation involving rotary adenosine triphosphate synthesis, chemiosmosis principle, proton pumps and electron transport chain. Biochem. Insights, 11: 1178626418818442. https://doi.org/10.1177/1178626418818442
Mansilla, N., Racca, S., Gras, D.E., Gonzalez, D.H., and Welchen, E., 2018. The complexity of mitochondrial complex iv: An update of cytochrome c oxidase biogenesis in plants. Int. J. mol. Sci., 19: 662. https://doi.org/10.3390/ijms19030662
Mordaunt, D.A., Jolley, A., Balasubramaniam, S., Thorburn, D.R., Mountford, H.S., Compton, A.G., Nicholl, J., Manton, N., Clark, D., and Bratkovic, D., 2015. Phenotypic variation of ttc19-deficient mitochondrial complex iii deficiency: A case report and literature review. Am. J. med. Genet. A, 167: 1330-1336. https://doi.org/10.1002/ajmg.a.36968
Nelson, D.L., and Cox, M.M., 2017. Lehninger’s principles of biochemistry. seventh edition. WH Freeman and Company.
Pal, A., Pal, A., Banerjee, S., Batabyal, S., and Chatterjee, P., 2019. Mutation in cytochrome b gene causes debility and adverse effects on health of sheep. Mitochondrion, 46: 393-404. https://doi.org/10.1016/j.mito.2018.10.003
Patel, M.R., Miriyala, G.K., Littleton, A.J., Yang, H., Trinh, K., Young, J.M., Kennedy, S.R., Yamashita, Y.M., Pallanck, L.J., and Malik, H.S., 2016. A mitochondrial DNA hypomorph of cytochrome oxidase specifically impairs male fertility in Drosophila melanogaster. Elife, 5: e16923. https://doi.org/10.7554/eLife.16923.001
Pradhan, M., Pal, A., Samanta, A., Banerjee, S., and Samanta, R., 2018. Mutations in cytochrome b gene effects female reproduction of ghungroo pig. Theriogenology, 119: 121-130. https://doi.org/10.1016/j.theriogenology.2018.05.015
Rahat, M.A., Haris, M., Ullah, Z., Ayaz, S.G., Nouman, M., Rasool, A., and Israr, M., 2020. Domestic animals’ identification using pcr-rflp analysis of cytochrome b gene. Adv. Life Sci., 7: 113-116.
Ramalho-Santos, J., Varum, S., Amaral, S., Mota, P.C., Sousa, A.P., and Amaral, A., 2009. Mitochondrial functionality in reproduction: From gonads and gametes to embryos and embryonic stem cells. Hum. Reprod. Update, 15: 553-572. https://doi.org/10.1093/humupd/dmp016
Ramsay, R.R., 2019. Electron carriers and energy conservation in mitochondrial respiration. Chem. Texts, 5: 1-14. https://doi.org/10.1007/s40828-019-0085-4
Reid, R., 2018. Applications of the mitochondrion in forensic DNA typing. In: DNA fingerprinting: Advancements and future endeavors. Springer, pp. 241-255. https://doi.org/10.1007/978-981-13-1583-1_14
Saikia, J., Roy, T., Naskar, S., Das, B., and Ferdoci, A., 2015. Molecular characterization of cytochrome b gene in indigenous pig. Indian J. Anim. Res., 49: 196-198. https://doi.org/10.5958/0976-0555.2015.00092.8
Sayers, E.W., Cavanaugh, M., Clark, K., Pruitt, K.D., Schoch, C.L., Sherry, S.T. and Karsch-Mizrachi, I., 2021. Genbank. Nucl. Acids Res., 49: D92-D96. https://doi.org/10.1093/nar/gkaa1023
Seddigh, S. and Darabi, M., 2018. Functional, structural, and phylogenetic analysis of mitochondrial cytochrome b (cytb) in insects. Mitochondrial DNA A, 29: 236-249. https://doi.org/10.1080/24701394.2016.1275596
Sievers, F., Wilm, A., Dineen, D., Gibson, T.J., Karplus, K., Li, W., Lopez, R., McWilliam, H., Remmert, M., and Söding, J., 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using clustal omega. Mol. Syst. Biol., 7: 539. https://doi.org/10.1038/msb.2011.75
Smith, J.L., Zhang, H., Yan, J., Kurisu, G. and Cramer, W.A., 2004. Cytochrome bc complexes: A common core of structure and function surrounded by diversity in the outlying provinces. Curr. Opin. Struct. Biol., 14: 432-439. https://doi.org/10.1016/j.sbi.2004.06.004
Stefely, J.A., and Pagliarini, D.J., 2017. Biochemistry of mitochondrial coenzyme q biosynthesis. Trends biochem. Sci., 42: 824-843. https://doi.org/10.1016/j.tibs.2017.06.008
Strand, D.D., Fisher, N., and Kramer, D.M., 2017. The higher plant plastid nad (p) h dehydrogenase-like complex (ndh) is a high efficiency proton pump that increases atp production by cyclic electron flow. J. biol. Chem., 292: 11850-11860. https://doi.org/10.1074/jbc.M116.770792
Studio, D., 2008. Discovery studio. Accelrys (2.1).
Tikhonov, A.N., 2018. The cytochrome b 6 f complex: Biophysical aspects of its functioning in chloroplasts. In: Membrane protein complexes: Structure and function. Springer. pp. 287-328. https://doi.org/10.3389/fpls.2013.00434
Tucker, E.J., Wanschers, B.F., Szklarczyk, R., Mountford, H.S., Wijeyeratne, X.W., van den Brand, M.A., Leenders, A.M., Rodenburg, R.J., Reljić, B., and Compton, A.G., 2013. Mutations in the uqcc1-interacting protein, uqcc2, cause human complex iii deficiency associated with perturbed cytochrome b protein expression. PLoS Genet., 9: e1004034. https://doi.org/10.1371/journal.pgen.1004034
Wanschers, B.F., Szklarczyk, R., van den Brand, M.A., Jonckheere, A., Suijskens, J., Smeets, R., Rodenburg, R.J., Stephan, K., Helland, I.B. and Elkamil, A., 2014. A mutation in the human cbp4 ortholog uqcc3 impairs complex iii assembly, activity and cytochrome b stability. Hum. mol. Genet., 23: 6356-6365. https://doi.org/10.1093/hmg/ddu357
Wolf, D.P., Hayama, T., and Mitalipov, S., 2017. Mitochondrial genome inheritance and replacement in the human germline. EMBO J., 36: 2177-2181. https://doi.org/10.15252/embj.201797606
Zhu, G., Zeng, H., Zhang, S., Juli, J., Pang, X., Hoffmann, J., Zhang, Y., Morgner, N., Zhu, Y., and Peng, G., 2020. A 3.3 å-resolution structure of hyperthermophilic respiratory complex iii reveals the mechanism of its thermal stability. Angew. Chem., 132: 351-359. https://doi.org/10.1002/ange.201911554
To share on other social networks, click on any share button. What are these?