Relationship Between GHRL Gene Polymorphisms and Growth Traits in Pigs
Relationship Between GHRL Gene Polymorphisms and Growth Traits in Pigs
Xin Xing Dong1*, Gai Cui Bai2, Ming Li Li1, Guo Xiang Lan1, Da Wei Yan1, Xiao Yi Wang1, Qiang Chen1 and Shao Xiong Lu1*
1Yunnan Agricultural University, College of Animal Science and Technology, Yunnan, Kunming
2Dehong Animal Husbandry Station, Yunnan, Dehong
ABSTRACT
Ghrelin (GHRL) is a gastric endocrine hormone, which has biological functions of regulation of appetite, body weight, gastrointestinal motility and growth hormone secretion. The aim of this study was to investigate the novel single nucleotide polymorphisms (SNPs) at GHRL gene in three different pig breeds, including Duroc, Yorkshire and Landrace, and the association of the novel SNPs with growth traits. PCR-SSCP and sequencing were used to detect the GHRL gene polymorphisms in the present study. Three SNPs were detected, one in the exon 1 (g.3734 G>T), which was synonymous, and two SNPs in intron 4 (g.7691 A>C and g.8523 T>C). The sites g.3734 G>T and g.7691 A>C had significantly impact on body weight at 21 days (BW21) and body weight at 70 days (BW70), the days when the pigs reached the target weights of 30 kg(D30) and 100 kg(D100), average daily gain between 30 kg and 100 kg (ADG), and backfat thickness of 100kg body weight (BFT) (P < 0.05 or P < 0.01), while did not contribute birth weight and days to 50 kg body weight(D50) (P > 0.05). At g.8523 T>C site, compared to TC genotype, the individuals with TT genotype in Duroc had greater D100 and BFT (P < 0.05 or P < 0.01), which had lower ADG between 30 kg and 100 kg (P < 0.01). In Yorkshire and Landrace, the polymorphism g.8523 T>C had no effect on the tested traits (P > 0.05). Our findings suggested that the 3 sites (g.3734 G>T, g.7691 A>C and g.8523 T>C) polymorphism in porcine GHRL gene might be one of the useful genetic marker in pig breeding and which needed to be further studied.
Article Information
Received 22 October 2019
Revised 05 December 2021
Accepted 22 December 2021
Available online 09 February 2022
(early access)
Published 22 October 2022
Authors’ Contribution
XXD and QC analyzed the data. XXD and SXL wrote the manuscript. GCB, MLL, GXL and DWY measured performance of pigs. XXD, GCB and MLL performed genotyping. DWY collected samples.
Key words
GHRL gene, Pig, Polymorphism, Growth traits, Meat productivities
DOI: https://dx.doi.org/10.17582/journal.pjz/20191022081026
* Corresponding author: 86127447@qq.com
0030-9923/2023/0001-145 $ 9.00/0
Copyright 2023 by the authors. Licensee Zoological Society of Pakistan.
This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
INTRODUCTION
Ghrelin is a 28 amino acid peptide produced primarily by the oxyntic cells of the stomach, which expressed the marked effects through binding to the endogenous ligand of the growth hormone secretagogue receptor (GHS-R) (Kojima et al., 1999). It requires an octanoyl group on the third amino acid for biological activity (Sun et al., 2011; Van et al., 2005) and involves in a series of biological functions including regulation of food intake, body weight (Wu et al., 2008; Carroll et al., 1998), hormone secretion (Hashizume et al., 2003; Rak et al., 2008), gastrointestinal (GI) motility (Kotunia et al., 2006), glucose release (Gaytan et al., 2004), cardiovascular functions (Hedayati et al., 2009), enzyme release (Rak et al., 2008), cell proliferation and reproduction in pigs through binding to GHS-R 1a or unidentified receptors (Rak et al., 2008).Many biological functions and their mechanisms of action have been implicated in other species such as human, fish and bird, but they are still unclear in pigs. Therefore, further research on this species needs to be performed in order to gain a better understanding of ghrelin peptide.
The pig GHRL gene is located in the q31 region of chromosome 13 and the cDNA sequence is about 2 kb, including 5 exons and 4 introns (Kojima et al., 1999). The GHRL gene is associated with growth traits and meat productivities in pigs, explicitly with lion-eye area, backfat thickness, residual feed intake, FCR and partial efficiency of growth (Tanaka et al., 2015; Ropkamolik et al., 2011; Wojtysiak et al., 2011; Kim et al., 2004).
The objectives of this study were to detect GHRL polymorphisms in terms of SNP and to test the impacts of polymorphisms on growth traits in three commercial pig breeds, including Duroc, Yorkshire and Landrace.
MATERIALS AND METHODS
Animals and trait measurements
Pigs (391) from 3 pure breeds (Yorkshire, 260; Landrace, 72; Duroc, 59) were used as the experimental populations in this study and only noninbred individuals for at least 3 generations were included. These pigs were randomly sampled in four batches and performance tested form born to 210 days of age under commercial conditions. During the test period, they had ad libitum access to commercial diets. The feeding program was based on two compound feeds: the first was for growers from 30 kg to 70 kg of body weight and the second feed was for finishers weighing from 70 to 100 kg. The energy, crude protein and digestible protein of grower diet was at least 13.5MJ/Kg, between 17-19% and at least 14.0, respectively. The energy, crude protein and digestible protein of finishers diet was at least 13.0 MJ/Kg, between 16-18% and at least 12.8 respectively. The growth traits included live body weight at birth (BW0), 21 days (BW21) and 70 days (BW70), backfat thickness (BFT), average daily gain (ADG) and the age when they reached the target weights (TW). BFT was recorded 5 cm off the midline at the position of the last rib by ultrasound when the pigs were 100kg (Piglog 105; SFK-Technology, Herlev, Denmark). The ADG was the rate of weight gain per day over the period between the pigs reached 30kg and 100kg. The TW were set at 30kg (D30), 70kg (D70) and 100kg (D100). All experimental procedures were performed according to the Guide for Animal Care and Use of Laboratory Animals in the Institutional Animal Care and Use Committee of Yunnan Agricultural University. The experimental protocol was approved by the Department Animal Ethics Committee of Yunnan Agricultural University.
Primers and genotype determination by PCR-SSCP
Genomic DNA was extracted from the ear tissues using a standard phenol-chloroform method then quantified using a NanoDrop spectrophotometer (GE Healthcare Life Sciences, Uppsala, Sweden). SNPs of the pig GHRL gene (GenBank, accession number NC_010455) were screened from the coding region and partial intron region. The primers used for SNP screening are shown in Table I. PCR amplifications were carried out in 25 μL reaction volumes containing at the following final concentrations: 50 ng of template DNA, 400 μM of dNTPs (Sangon, China), 0.25 μM of each primer and 1 unit (U) of Taq polymerase (TaKaRa, Japan). The PCR protocol consisted of an initial denaturation at 94 °C for 5 min, followed by 36 cycles of denaturation at 94 °C for 30 s, annealing at 60°C for 30 s, and extension at 72 °C for 30 s, with a final extension at 72 °C for 10 min. The PCR products were sequenced on both strands. The sequences obtained were compared with MEGA v4.0 (www.megasoftware.net/). The PCR-SSCP method was used to scan mutation within the amplified region. The detected SNPs were genotyped by using PCR-SSCP assays. A 5 μL aliquot of each amplicon was diluted in denaturing solution (98% formamide, 10 mM EDTA, 0.025% bromophenol blue, 0.025% xylene-cyanol) denatured at 95 °C for 5 min, rapidly cooled on ice and resolved in acrylamide: Bisacrylamide (29:1) (Bio-Rad) gels at 400 V for 4 h at 4 °C, in 1 × TBE buffer (89 mM Tris base, 89 mM boric acid, 2 mM EDTA, pH 8.0). The gels were silver-stained according to the method of (Bassam and Gresshoff, 2007). After electrophoresis, the PCR products of different electrophoresis patterns were sequenced and polymorphism of the GHRL gene was studied.
Statistical analysis
Gene frequencies were determined by direct counting. Gene heterozygosity (H) and polymorphism information content (PIC) were calculated according to Nei’s and Botstein’s methods (Nei and Roychoudhury, 1974; Botstein et al., 1980), respectively. The formulas were as follows:
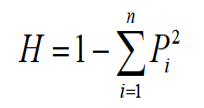
Table I. Segmental amplification primers of GHRL gene.
|
No. of primer |
Sequence of primers (5’→3’) |
Location |
Size of products |
|
1 |
Forward primer: ACCCACGCCAACCCAGCAAT |
exon1, part intron1 3556-3921 bp |
366 |
|
Reverse primer: GCAAGGTCCCAGGAGACAAGG |
|||
|
2 |
Forward primer: TGGAACAGGTGCTTTAGTCGGC |
part intron1, exon2 4066-4376 bp |
311 |
|
Reverse primer: TGGAAGGGCTGGCATGAGGT |
|||
|
3 |
Forward primer: CCCATACCTTGCTCTGCTT |
part intron2, exon3 4450-4711 bp |
262 |
|
Reverse primer: ACCCTCCACTCCCTCATCT |
|||
|
4 |
Forward primer: ATAGCAGGGGCAGGAGGGTGT |
part intron3, exon4, part intron 4 7535-7792 bp |
260 |
|
Reverse primer: AGGCAGGGATGGGGAAGGAAA |
|||
|
5 |
Forward primer: AGCCACAGCAGTGACAACC |
part intron4, exon5 8446-8816 bp |
371 |
|
Reverse primer: CCTGACCGTAGACCAAGAATG |
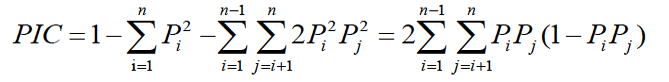
(Pi, Pj are the frequency of the i and j allele, n is the number of allele).
The genotype and allele frequencies were tested for Hardy-Weinberg equilibrium using the χ2 test. The association analyses between single SNP and measured traits were performed with a statistical model using the MIXED model procedures (SAS10; SAS Institute, Inc.). The following model was used:
yijk= μ + Gi + Sj + eijk
Where yijk is the observation of the trait; μ is the population mean; Gi is the fixed effect of the genotype; Sj is the fixed effect of sex; eijk is the random residual error. Least square means and respective standard errors for genotypes were estimated based on the statistical model and significant levels were set at P < 0.05 and P < 0.01, respectively.
RESULTS
Polymorphisms and sequence variation of GHRL gene
In present study, the five fragments were amplified from genomic DNA. Thereafter, three unique SSCP patterns in the 366 bp, 260 bp and 371bp PCR product were detected after PCR-SSCP analysis respectively (Figs. 1, 2, 3). DNA sequencing analysis showed that, one SNP within the coding sequence of GHRL, located in the exon 1 (g.3734 G>T), and two SNPs in intron 4 (g.7691 A>C and g.8523 T>C). However, the exonic SNP was synonymous.
Genotype frequencies
Genotype and allele frequencies of g.3734 G>T, g.7691 A>C and g.8523 T>C within the three breeds is shown in Table II. Middle genetic diversity of GHRL gene was revealed from the values of PIC. The g.3734 G>T polymorphism are showed the similar pattern in the studied groups, which means that the frequency of the G allele was always greater than that of the T allele. The frequency of the GG homozygote was higher than the TT genotype, and the frequency of the GT heterozygote was in the middle. The breeds were at Hardy-Weinberg equilibrium with respect to this marker. The heterozygosity (H) of the mutant loci was 0.4677, 0.4297 and 0.4200 respectively in the three pig breeds.
At the g.7691 A>C site, the frequency of the A allele was higher than the C allele and the AA genotype was higher than AC, CC genotypes in the three breeds. The genetic diversity of this site was at a moderate polymorphism, and the heterozygosity of Yorkshire (0.4638) was higher than that of Duroc (0.3240) and Landrace (0.3133). The SNP deviated from Hardy-Weinberg equilibrium in the three pig populations (P<0.01).
At the g.8523 T>C site, the frequency of TC genotype was the highest in Duroc and Landrace pigs (0.6441 and 0.6111, respectively). Genotype CC was not found in Duroc. The frequency of the T allele was higher than the C allele in the three breeds.
Based on the χ2 statistics, the breeds were not at Hardy-Weinberg equilibrium with respect to this marker (P<0.01).
Relationship between GHRL gene genotypes and recorded traits
The results of the association analyses of GHRL gene genotypes and the growth traits within the three different populations are shown in Table III.
In the Duroc breed, individuals with TT genotype of G3734T had higher BW70 than those with the GT genotype (P<0.05). However, heterozygote GT showed significantly higher D30 than the homozygote TT (P<0.01). In the Landrace breed, individuals with TT genotype at G3734T had larger BW21, BW70, D100, BFT and less ADG than those with the other two genotypes. In the Yorkshire breed, individuals with GT genotype had larger BW70, than those with TT genotype (P<0.05). The TT genotype had significantly larger D100 and BFT than those with the CT and TT genotype (P<0.01 for D100 and P<0.05 for BFT).
At the g.7691 A>C site, individuals with AA and AC genotypes had higher BW21 than those with the CC genotype (P<0.05) in the Duroc breed, however the two genotypes were associated with higher BW70 in the Yorkshire breed. In the Duroc breed, individuals with AC genotype reached the TW of 30kg and 100 kg at 69.15 days and 161.05 days and were about 4 (for D30) and 7 days (for D100) shorter than individuals with the other two genotypes (P<0.05). Interestingly, in the Yorkshire breed, CC genotype reduced BW70 and slowed the D30, but it shortened D100 and benefited ADG.
At the g.8523 T>C site, Duroc pigs with TT genotype had slower ADG and consequently lengthened the days reached TW 100kg, but thicker BFT was also found for the genotype No significant difference was found for Landrace and Yorkshire breed.
Table II. The genotypic and allelic frequencies of SNPs in GHRL gene in three pig breeds.
|
SNP site |
Breed |
Genotype |
Genotype frequency |
Allele |
Allele frequency |
χ2 |
H |
PIC |
|
g.3734 G>T |
D |
GG |
0.4576(27) |
G |
0.6271 |
4.4679* |
0.4677 |
0.3583 |
|
GT |
0.3390(20) |
|||||||
|
TT |
0.2034(12) |
T |
0.3729 |
|||||
|
L |
GG |
0.4722(34) |
G |
0.6875 |
0.0003 |
0.4297 |
0.3374 |
|
|
GT |
0.4306(31) |
|||||||
|
TT |
0.0972(7) |
T |
0.3125 |
|||||
|
Y |
GG |
0.5192(135) |
G |
0.7000 |
5.0375* |
0.4200 |
0.3318 |
|
|
GT |
0.3615(94) |
|||||||
|
TT |
0.1192(31) |
T |
0.3000 |
|||||
|
g.7691 A>C |
D |
AA |
0.7627(45) |
A |
0.7966 |
36.8947** |
0.3240 |
0.2715 |
|
AC |
0.0678 (4) |
|||||||
|
CC |
0.1695(10) |
C |
0.2034 |
|||||
|
L |
AA |
0.7361(53) |
A |
0.8056 |
22.3099** |
0.3133 |
0.2642 |
|
|
AC |
0.1389(10) |
|||||||
|
CC |
0.1250(9) |
C |
0.1944 |
|||||
|
Y |
AA |
0.5154(134) |
A |
0.6385 |
62.5092** |
0.4589 |
0.3577 |
|
|
AC |
0.2385 (62) |
|||||||
|
CC |
0.2462(64) |
C |
0.3615 |
|||||
|
g.8523 T>C |
D |
TT |
0.3559(21) |
T |
0.6780 |
13.3119** |
0.4367 |
0.3413 |
|
TC |
0.6441(38) |
|||||||
|
CC |
0.0000(0) |
C |
0.3220 |
|||||
|
L |
TT |
0.3750(27) |
T |
0.6806 |
11.8391** |
0.4348 |
0.3403 |
|
|
TC |
0.6111(44) |
|||||||
|
CC |
0.0139(1) |
C |
0.3194 |
|||||
|
Y |
TT |
0.5115(133) |
T |
0.7462 |
14.8111** |
0.3788 |
0.3071 |
|
|
TC |
0.4692(122) |
|||||||
|
CC |
0.0192(5) |
C |
0.2538 |
Note: D, Duroc; L, Landrace; Y, Yorkshire; 3.84 < χ2 < 6.63 represents P < 0.05; χ2 > 6.63 represents P < 0.01. * P < 0.05; ** P < 0.01.
Table III. Least squares means and standard error of genotypes of three SNP in GHRL gene for growth traits.
|
Site |
Breed |
Genotype |
# Animal |
BW0 |
BW21 |
BW70 |
D30 |
D50 |
D100 |
ADG |
BFT |
|
G3734T |
D |
GG |
27 |
1.87± 0.07 |
7.59± 0.13 |
28.13± 0.40AB |
72.77± 0.68AB |
101.60± 0.73 |
167.37± 0.99 |
743±9 |
9.20± 0.21 |
|
GT |
20 |
1.86± 0.07 |
7.45± 0.14 |
27.04± 0.45B |
74.50± 0.75A |
101.78± 0.81 |
169.01± 1.10 |
746±10 |
9.54± 0.23 |
||
|
TT |
12 |
1.77± 0.09 |
7.55± 0.17 |
29.17± 0.61A |
70.73± 1.02B |
101.16± 1.10 |
167.12± 1.50 |
728±13 |
9.38± 0.31 |
||
|
L |
GG |
34 |
1.74± 0.04 |
6.84± 0.16b |
26.95± 0.46b |
73.97 ±0.80 |
102.99± 0.79 |
168.57 ±0.85b |
743±9A |
9.55± 0.18b |
|
|
GT |
31 |
1.73± 0.03 |
7.32± 0.13b |
27.74± 0.46ab |
73.27± 0.80 |
102.25± 0.79 |
167.59± 0.86b |
745±9A |
9.02± 0.18c |
||
|
TT |
7 |
1.63± 0.08 |
7.70± 0.32a |
29.69± 0.99a |
70.62± 1.71 |
101.99± 1.70 |
172.44± 1.83a |
691±18B |
10.04± 0.39a |
||
|
Y |
GG |
135 |
1.72± 0.03 |
7.22± 0.11 |
27.00± 0.23ab |
73.61± 0.42 |
102.34± 0.44 |
169.65± 0.46B |
736±5 |
9.44± 0.09b |
|
|
GT |
94 |
1.73± 0.02 |
7.30± 0.09 |
27.24± 0.26a |
73.56± 0.48 |
101.90± 0.50 |
169.84± 0.53B |
732±5 |
9.54± 0.11ab |
||
|
TT |
31 |
1.71± 0.04 |
7.16± 0.19 |
26.19± 0.46b |
74.69± 0.86 |
101.98± 0.89 |
172.75± 0.94A |
718±9 |
9.96± 0.19a |
||
|
A7691C |
D |
AA |
45 |
1.84± 0.05 |
7.62± 0.09a |
27.87± 0.33 |
73.30± 0.55a |
101.61± 0.57 |
168.57± 0.73a |
737±7 |
9.26± 0.16 |
|
AC |
4 |
2.08± 0.24 |
7.71± 0.43a |
29.58± 1.04 |
69.15± 1.73b |
100.20± 1.81 |
161.05± 2.30b |
763±22 |
9.06± 0.50 |
||
|
CC |
10 |
1.85± 0.17 |
6.79± 0.31b |
27.36± 0.66 |
73.66± 1.09a |
102.06± 1.14 |
168.23± 1.46a |
748±14 |
9.92± 0.32 |
||
|
L |
AA |
53 |
1.73± 0.03 |
7.01± 0.14 |
27.31± 0.39 |
73.63± 0.65 |
102.63± 0.64 |
168.20± 0.72 |
744±7 |
9.18± 0.15B |
|
|
AC |
10 |
1.75 ±0.05 |
7.29± 0.21 |
28.82± 0.85 |
71.43± 1.43 |
101.56± 1.41 |
169.11± 1.58 |
718±16 |
10.13± 0.33A |
||
|
CC |
9 |
1.68± 0.06 |
7.48± 0.25 |
27.47± 0.91 |
74.07± 1.54 |
103.22± 1.51 |
168.71± 1.70 |
742±17 |
9.61± 0.35AB |
||
|
Y |
AA |
134 |
1.70± 0.02 |
7.23± 0.09 |
27.23± 0.22a |
73.23± 0.41b |
101.64± 0.43 |
170.85± 0.45A |
721±4B |
9.48± 0.09 |
|
|
AC |
62 |
1.75± 0.03 |
7.34± 0.13 |
27.22± 0.32a |
73.45± 0.60ab |
102.89± 0.63 |
170.50± 0.66A |
728±6B |
9.52± 0.13 |
||
|
CC |
64 |
1.77± 0.03 |
7.24± 0.14 |
26.33± 0.32b |
74.64± 0.58a |
102.44± 0.61 |
168.08± 0.64B |
758±6A |
9.68± 0.13 |
||
|
T8523C |
D |
TT |
21 |
/ |
/ |
28.04± 0.47 |
72.68 ±0.78 |
101.72± 0.79 |
169.75± 1.04a |
725±9B |
9.96± 0.20A |
|
TC |
38 |
1.85 ±0.04 |
7.53± 0.09 |
27.82± 0.36 |
73.29± 0.61 |
101.51± 0.61 |
166.88± 0.81b |
751±7A |
9.01± 0.16B |
||
|
L |
TT |
27 |
/ |
/ |
27.28± 0.52 |
73.39± 0.89 |
103.64± 0.84 |
170.72± 0.88 |
723±9 |
9.57± 0.20 |
|
|
TC |
44 |
1.72± 0.03 |
7.15± 0.12 |
27.63± 0.44 |
73.48± 0.74 |
101.91± 0.70 |
166.86± 0.73 |
752±8 |
9.15± 0.16 |
||
|
CC |
1 |
1.81 |
8.06 |
27.92 |
72.21 |
100.89 |
165.67 |
750 |
11.57 |
||
|
Y |
TT |
133 |
/ |
/ |
27.04± 0.22 |
73.46± 0.41 |
101.59± 0.42 |
171.41± 0.44 |
719±4 |
9.67± 0.09 |
|
|
TC |
122 |
1.73± 0.02 |
7.25± 0.07 |
26.96± 0.24 |
74.00± 0.44 |
102.69± 0.45 |
168.42± 0.47 |
748±5 |
9.37± 1.00 |
||
|
CC |
5 |
1.60± 0.10 |
7.62± 0.45 |
26.92± 1.12 |
73.73± 2.08 |
104.05± 2.14 |
171.48± 2.22 |
719±22 |
9.89± 0.46 |
Note: A, B, estimates for each SNP and each trait with different superscripts differ (P < 0.01). a, b, estimates for each SNP and each trait with different superscripts differ (P < 0.05). # animal, indicates the number of animals. BW21, BW at 21 d of age; BW70, BW at 70 d of age. D30, days to 30 kg body weight; D50, days to 50 kg body weight, D100, days to 100 kg body weight. BFT, backfat thickness of 100kg body weight.
DISCUSSION
In previous studies, only one RFLP-BsrI polymorphism in the GHRL gene that was identified in silico mining porcine sequences was reported to be associated with BFT, marbling, and other carcass traits in a Berkshire and Yorkshire pigs (Kim et al., 2004). The BsrI polymorphism at the ghrelin gene locus is potentially associated with carcass and meat quality traits in Polish Landrace pigs and could be considered as one of meat quality markers in the future (Fang et al., 2011; Wojtysiak et al., 2011). However, instead of BsrI polymorphism reported in previous studies, three novel SNPs were detected in GHRL gene in our study, including one SNP located in the exon 1 (g.3734 G>T), and two SNPs in intron 4 (g.7691 A>C and g.8523 T>C), which were associated with growth traits in different breeds.
Although the exonic SNP (g.3734 G>T) was synonymous, it is also associated with BW21, BW70, D30, D100, ADG and BFT. Additionally, not only nonsynonymous mutations that alter amino acid a protein polypeptide chain are functional in gene expression process, but also synonymous mutations that located in introns or 5′ UTR regions may determine mRNA stability or affect transcriptional efficiency of numerous genes (Greenwood et al., 2003; Stefanovic et al., 1999).
Ghrelin can reduce anorexia time during weaning and increase average daily gain. Moreover, it is potentially able to improve resistance to pathological and environmental challenges during weaning, and then shortens the days that required to reach the slaughter weights (Carroll et al., 1998; Scerif et al., 2011). Analyses of the relationships between genotypes of the three SNPs at g.3734 G>T, g.7691 A>C and g.8523 T>C and body weight showed different patterns in the three breeds. For example, at the g.3734 G>T site, TT genotypes increased BW70 for Duroc as well as BW21 and BW70 for Landrace pigs, but decreased BW70 for Yorkshire breed. Nevertheless, the homozygote TT linked with thicker BFT and longer D100 in Landrace and Yorkshire populations. At the g.7691 A>C site, overdominances of heterozygotes was significantly different from that of homozygotes, which has attracted the attention, e.g., BW21, D30 and D100 in Duroc and BFT in Landrace. However, no overdominance presented at the A7691C site in Yorkshire pigs, because the extreme least square means were only observed for homozygotes. Basically, AA genotypes benefited growth in early life and CC genotypes favored overall growth as the AA genotype was associated with heavier BW at 70 days of age and shorter days to reach TW of 30kg, while the CC genotype increased ADG and consequently shortened the raising periods required from birth to market weight. At the g.8523 T>C site, different genotypes had influence on D100, ADG of 30-100 kg and BFT of 100kg body weight in the Duroc breed. However, the genotype difference did not significantly affect growth traits in the Yorkshire breed.
Back fat thickness is a measure of carcass fatness and can be easily obtained after slaughtering or in vivo by ultrasound measurement. Therefore, this phenotype is included as a target trait in breeding programs in purebred and commercial lines. Several human obesity-related genes have been already investigated in pigs with the objective to identify DNA markers associated with fat deposition traits (Switonski et al., 2010; Dantas et al., 2011). The GHRL gene was implicated in playing a role in backfat thickness in Italian Large White (Jina et al., 2011; Fontanesi et al., 2012). In this research, all SNPs detected in GHRL were associated with BFT in the Landrace. However, only one significant SNP was found for Duroc and Yorkshire breeds. At the g.8523 T>C site, individuals with TT genotype had higher BFT of 100kg body weight than those of the TC genotype in the Duroc breed. In the Yorkshire breed, individuals with TT genotype of G3734T had larger BW70, than those with TT genotype (P<0.05).
Our findings suggested polymorphism in GHRL gene might be one of the important genetic factors influencing growth and backfat thickness in the three pig breeds. Although the mechanisms involved in the association of alternative genotypes in the intronic SNP were not understood, the important role of the non-coding portion of genomes was widely acknowledged. Mutations within introns could affect the splice donor, acceptor site or regions nearby and regulatory motifs within introns (Cheong et al., 2006). So, two mutations in intron 4 of the GHRL gene might influence body weight and backfat thickness in pig. The significant effects of GHRL genotypes on body weight presented in this study provided a basis for further study of the function of the GHRL gene in pigs, and show some possibilities for the application of the GHRL gene as a molecular marker in commercial breeding. Moreover, additional research with a bigger population should be carried out in further studies.
ACKNOWLEDGEMENTS
This work was financially supported by the Leading Talents of Industrial Technology of Yunnan Province (YNWR-CYJS-2018-056), Yunnan University Science and technology innovation team.
Statement of conflict of interest
The authors have declared no conflict of interest.
REFERENCES
Bassam, B.J. and Gresshoff, P.M., 2007. Silver staining DNA in polyacrylamide gels. Nature Prot., 2: 2640-2654. https://doi. org/ 10.1038/nprot.2007.330.
Botstein, D., White, R.L., Skolnick, M., Davis, R.W., 1980. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet., 32: 314-331.
Carroll, J.A., Veum, T.L., and Matteri, R.L., 1998. Endocrine responses to weaning and changes in post-weaning diet in the young pig. Domest. Anim. Endocrinol., 15: 183–194. https://doi.org/10.1016/S0739-7240(98)00006-X
Cheong, H.S., Yoon, D.H., Kim, L.H., Park, B.L., Choi, Y.H., Chung, E.R., Cho, Y.M., Park, E.W., Cheong, I.C., Oh, S.J., Yi, S.G., Park, T., and Shin, H.D., 2006. Growth hormone-releasing hormone (GHRH) polymorphisms associated with carcass traits of meat in Korean cattle. BMC Genet., 7: 35. https://doi.org/10.1186/1471-2156-7-35
Dantas, V.G.L., Furtado-Alle, L., Souza, Ricardo. L.R., Souza, Eleidi, A., and Chautard F.M., 2011. Obesity and variants of the GHRL (ghrelin) and BCHE (butyrylcholinesterase) genes. Genet. Mol. Biol., 34: 205–207. https://doi.org/10.1590/S1415-47572011000200006
Fang, M.X., Ying, L.I., Hai-Ping, X.U., Xie, L., Liao, X.D., and Liang, M., 2011. Associations of ghrelin(ghrl) and its receptor(ghsr) genes polymorphisms with duck growth and carcass traits. Chinese J. Anim. Vet. Sci., 42: 18-24.
Gaytan, F., Barreiro, M.L., Caminos, J.E., Chopin, L.K., Herington, A.C., and Morales, C., 2004. Expression of ghrelin and its functional receptor, the type 1a growth hormone secretagogue receptor, in normal human testis and testicular tumors. J. clin. Endocrinol. Metab., 89: 400-409. https://doi.org/10.1210/jc.2003-031375
Greenwood, T.A., and Kelsoe, J.R., 2003. Promoter and intronic variants affect the transcriptional regulation of the human dopamine transporter gene. Genomics, 82: 511-520. https://doi.org/10.1016/S0888-7543(03)00142-3
Hashizume, T., Horiuchi, M., Tate, N., Nonaka, S., Mika, U., and Kojima, M., 2003. Effects of ghrelin on growth hormone secretion from cultured adenohypophyseal cells in pigs. Domest. Anim. Endocrinol., 24: 209-218. https://doi.org/10.1016/S0739-7240(02)00240-0
Hedayati, N., Annambhotla, S., Jiang, J., Wang, X., Chai, H., and Lin, PH., 2009. Growth hormone-releasing peptide ghrelin inhibits homocysteine-induced endothelial dysfunction in porcine coronary arteries and human endothelial cells. J. Vasc. Surg., 49: 199-207. https://doi.org/10.1016/j.jvs.2008.08.065
Jina, Q.J., Fang, X.T., Zhang, C.L., Yang, L., Sun, J.J., Chen, D.X., Shi, X.Y., Du, Y., Lan, X.Y., and Chen., H., 2010. A novel SNP of the GHRL gene in goat and its association with growth traits. Small Rumin. Res., 90: 150-152. https://doi.org/10.1016/j.smallrumres.2010.01.001
Kim, K.S., Thomen, H., and Bastiaansen, J., 2004. Investigation of obesity candidate genes on porcine fat deposition quantitative trait loci regions. Obes. Res., 12: 1981-1994. https://doi.org/10.1038/oby.2004.249
Kojima, M., Hosoda, H., Date, Y., Nakazato, N., Matsui, H., and Kangawa, K., 1999. Ghrelin is a growth hormone-releasing acylated peptide from stomach. Nature, 402: 656-660. https://doi.org/10.1038/45230
Kotunia, A., and Zabielski, R., 2006. Ghrelin in the postnatal development of the gastrointestinal tract. J. Physiol. Pharmacol., 57: 97-111.
Nei, M. and Roychoudhury, A.K.J., 1974. Sampling variances of heterozygosit and genetic distance. Genetics, 76: 379-390. https://doi.org/10.1093/genetics/76.2.379
Rak, A., and Gregoraszczuk, E.L., 2008. Local feedback loop of ghrelin-GH in the pig ovary: Action on estradiol secretion, aromatase activity and cell apoptosis. Growth Horm. IGF Res., 18: 221-227. https://doi.org/10.1016/j.ghir.2007.09.004
Ropkamolik, K., Oczkowicz, M., Piórkowska, K., and Różycki, M., 2011. New polymorphisms and expression of the porcine ghrelin (ghrl) gene in different pig breeds. J. Anim. Feed Sci., 20: 186-191. https://doi.org/10.22358/jafs/66170/2011
Scerif, M., Goldstone, A.P., and Korbonits, M., 2011. Ghrelin in obesity and endocrine diseases. Mol. Cell. Endocrinol., 340: 15-25. https://doi.org/10.1016/j.mce.2011.02.011
Stefanovic, B., Hellerbrand, C., and Brenner, D. A., 1999. Regulatory role of the conserved stem-loop structure at the 5′ end of collagen α1 (I) mRNA. Mol. Cell. Biol., 19: 4334-4342. https://doi.org/10.1128/MCB.19.6.4334
Sun, J., Jin, Q., Zhang, C., Fang, X., Gu, C., Lei, C., Wang, J., and Chen, H., 2011. Polymorphisms in the bovine ghrelin precursor (ghrl) and syndecan-1 (sdc1) genes that are associated with growth traits in cattle. Mol. Biol. Rep., 38: 3153-3160. https://doi.org/10.1007/s11033-010-9986-4
Switonski, M., Stachowiak, M., Cieslak, J., Bartz, M., and Grzes, M., 2010. Genetics of fat tissue accumulation in pigs: A comparative approach. J. appl. Genet., 51: 153-168. https://doi.org/10.1007/BF03195724
Tanaka, K., Takizawa, T., Oki, O., Fukawa, K., Ito, T., Miyabe, M., Mannen, H., Kurosawa, Y., and Hirose, K., 2015. Allele frequency for c.335a>c polymorphisms in porcine ghrelin/obestatin prepropeptide gene and association analysis with performance traits in various pig breeds. Czech J. Anim. Sci., 60: 411-416. https://doi.org/10.17221/8459-CJAS
Van der Lely, A.J., Tschop, M., Heiman, M.L., and Ghizo, E., 2005. Biological, physiological, pathophysiological and pharmacological aspects of ghrelin. Endocr. Rev., 25: 426-457. https://doi.org/10.1210/er.2002-0029
Wojtysiak, D., and Kaczor, U., 2011. Effect of polymorphisms at the ghrelin gene locus on carcass, microstructure and physicochemical properties of longissimus lumborum muscle of Polish Landrace pigs. Meat Sci., 89: 514-518. https://doi.org/10.1016/j.meatsci.2011.05.019
Wu, X., Tang, M., Ma, Q., Hu, X., and Ji, C., 2008. Effects of exogenous ghrelin on the behaviors and performance of weanling piglets. Asian-Australas. J. Anim. Sci., 21: 861-867. https://doi.org/10.5713/ajas.2008.70574
To share on other social networks, click on any share button. What are these?










