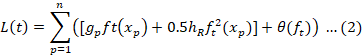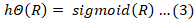Investigation of Bovine Disease and Events through Machine Learning Models
Research Article
Investigation of Bovine Disease and Events through Machine Learning Models
Ghalib Nadeem* and Muhammad Irfan Anis
High Performance Research Group, FEST, Iqra University Defence View, Karachi, Pakistan.
Abstract | Bovine disease identification utilizing multiple information sources and methods is widely applicate in the field of bovine disease prevention and health monitoring. Bovine disease detection is an emerging subject today to accomplish the farms demands of individuals across the globe. This research delves into the realm of bovine disease and event detection using advanced Machine Learning (ML) techniques. Focusing on the critical events of estrus, acidosis, mastitis, lameness, and calving, our study aims to revolutionize disease identification and timely intervention within the dairy industry. By leveraging four distinct ML models—Random Forest, XGBoost, Logistic Regression, and Single Perceptron we meticulously analyze four diverse data-sets to uncover intricate patterns and unveil hidden insights. The efficiency of random sampling in resolving the class imbalance issue is tested along with the validity and adaptability of these models utilizing GridSearchCV optimum parameter modification. The performance evaluation is based on accuracy, precision, recall, F1 score, and Area Under the Curve (AUC) metrics. With a resounding highest accuracy metric, the Random Forest model achieves a notable accuracy of up to 98.25%, while the recall score of 100, and Precision up to 97% affirming its supremacy in classifying bovine events. This achievement underscores the efficacy of employing ML algorithms for accurate and timely disease identification. This ground-breaking fusion of ML techniques with bovine disease detection holds trans-formative potential, promising to elevate animal welfare standards, optimize dairy productivity, and usher in a new era of data-driven dairy management.
Received | October 25, 2023; Accepted | April 29, 2024; Published | May 10, 2024
*Correspondence | Ghalib Nadeem, High Performance Research Group, FEST, Iqra University Defence View, Karachi, Pakistan; Email: ghalibnadeem@iqra.edu.pk
Citation | Nadeem, G. and M.I. Anis. 2024. Investigation of bovine disease and events through machine learning models. Pakistan Journal of Agricultural Research, 37(2): 102-114.
DOI | https://dx.doi.org/10.17582/journal.pjar/2024/37.2.102.114
Keywords | Dairy industry, Estrus prediction, Machine learning, Random forest
Copyright: 2024 by the authors. Licensee ResearchersLinks Ltd, England, UK.
This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Introduction
The dairy industry plays a pivotal role in global food security by providing essential nutrition through milk production, relies heavily on the health and productivity of dairy cows. However, ensuring the health and well-being of dairy cows is of paramount importance for sustaining a thriving dairy industry, making bovine diseases a top concern for farmers worldwide. Bovine diseases, such as calving complications, mastitis, estrus, acidosis, and lameness, pose significant challenges. Detecting and diagnosing these diseases early is critical, as it enables timely intervention and effective disease management. Early detection not only facilitates prompt treatment but also minimizes the economic losses associated with reduced productivity and increased veterinary costs. Moreover, it prevents the spread of diseases within herds, safeguarding the overall dairy herd health. The potential consequences of undetected diseases can be far-reaching, impacting not only individual farms but also the broader dairy industry and global food supply chain.
Thus, the development of accurate and efficient bovine disease detection systems, especially utilizing machine learning models, holds immense promise for enhancing animal welfare, optimizing dairy productivity, and ensuring the sustainability and profitability of the dairy industry on a global scale. Machine learning in bovine disease detection offers a trans-formative approach to dairy health management. ML’s ability to detect diseases early, provide non-invasive and objective assessments, handle complex data patterns, and enable real-time monitoring and automation significantly enhances the overall efficiency, effectiveness, and sustainability of dairy farming practices. Researchers have built multiple ML-based bovine disease and event detection systems with various aspect of methods and techniques that can identify mastitis (Ghafoor et al., 2021), lameness (Post et al., 2020), calving (Santos et al., 2022), estrus (Wang et al., 2020), and acidosis (Wagner et al., 2020).
The objective is to offers a significant contribution to the field of agricultural informatics and bovine health management by the development of system that can specifically focusing on calving, mastitis, estrus, acidosis, and lameness. By harnessing the potential of machine learning and artificial intelligence, we present an innovative approach to improve disease detection accuracy, reduce economic losses, and enhance overall dairy productivity. A crucial phase of the dairy cow’s reproductive cycle is calving. For the health and safety of both the cow and the calf, prompt calving identification is essential. Dystocia and postpartum disorders can be avoided with prompt detection of calving issues, lowering the risk of mortality and improving total breeding success (Roche et al., 2023). Researchers have explored various techniques and methods to achieve this goal. Video analysis and image processing techniques have been employed to capture cow behavior during calving (Hyodo et al., 2023). Acoustic monitoring has been utilized to detect distinct vocal patterns associated with calving (Alexandra et al., 2020). Sensor-based monitoring, including wearable accelerometers, provides insights into cow movements and posture (Riaboff et al., 2022). Thermal cameras and infrared sensors have been used to measure body temperature changes, indicative of impending calving (Cantor et al., 2022). Additionally, bio-metric and physiological data analysis, such as heart rate and rumination patterns, offers valuable indicators (Alipio et al., 2022). Contextual data integration, real-time processing, and ensemble methods have also been explored for enhanced accuracy (García et al., 2020; Das et al., 2023; Ali et al., 2020).
Female cows go through a natural phase of sexual receptivity called estrus, which is important for the reproductive cycle. For optimal breeding management, optimum reproduction, and the highest possible breeding success rates, accurate estrus detection is necessary. Researchers have employed diverse techniques and methods to achieve early estrus detection. Acoustic monitoring has also been explored, capturing vocalizations and sounds indicative of estrus (Wang et al., 2022). Contextual data integration, involving the fusion of information from multiple sensors, has shown promise in improving detection performance (Leliveld et al., 2021; Huang et al., 2023). Sensor-based monitoring using accelerometers and motion sensors has provided valuable insights into cow activity patterns during estrus (Arcidiacono et al., 2020).
In dairy cattle, lameness is a serious economic and welfare concern. It limits the cow’s mobility, inflicts pain, and is frequently brought on by foot and leg injuries (Kang et al., 2022). Lameness raises the risk of other health problems and lowers productivity. Early detection of lameness is crucial for prompt treatment and preventing further complications. Researchers have explored diverse techniques and methods, harnessing the power of machine learning to achieve early lameness detection. Video analysis has also been employed, capturing cow behavior and locomotion, Machine learning algorithms have been trained on this sensor data to accurately classify normal and lame gait patterns to identify lameness indicators (Van Nuffel et al., 2015). Wearable sensor-based monitoring, such as hoof-mounted accelerometers, provides real-time data on cow movement and activity, enabling the identification of lameness indicators (Haladjian et al., 2018; O’Leary et al., 2020). Additionally, data fusion techniques, combining information from multiple sources like video and sensor data, have been explored to enhance the accuracy of lameness detection (Zheng et al., 2023).
Mastitis is an infection-related mammary gland inflammation that frequently results from bacteria. It is a common and expensive condition that affects dairy cattle and causes decreased milk production and poor milk quality. For prompt intervention, the reduction of financial losses, and the preservation of milk yield and quality, accurate mastitis identification is essential. Researchers have explored a range of machine learning techniques to develop innovative methods for mastitis detection in bovine. In recent studies, ultrasound imaging has been used to assess udder health and identify early signs of mastitis (Themistokleous et al., 2023). Additionally, researchers have integrated data from milk composition analysis, such as somatic cell count and milk conductivity, into machine learning algorithms for mastitis prediction (Matera et al., 2022).
Due to a high-energy diet and an unbalanced rumen pH, dairy cows may develop acidosis, a metabolic condition. Reduced feed intake, subpar milk output, and other health issues follow. Acidosis can be managed promptly, preventing serious health problems and enhancing overall animal welfare. Researchers have embraced advanced machine learning techniques to develop cutting-edge methods for acidosis detection in bovine. Continuous rumen pH monitoring using wireless pH sensors has been explored to provide real-time insights into rumen health and acidosis risk (Han et al., 2022). Analyzing changes in rumination behavior allows for early identification of acidotic conditions. Additionally, data from feed intake monitoring systems have been incorporated into machine learning algorithms to further improve acidosis prediction (Wagner et al., 2020; Heirbaut et al., 2022).
Machine learning has revolutionized bovine event monitoring, providing real-time automated surveillance to alert farmers to critical situations. This technology enhances accuracy through early disease and event detection, ensuring non-invasive, stress-free monitoring. It eliminates human subjectivity and bias, resulting in more reliable diagnoses. This advancement also fosters cost-effectiveness and sustainable farming by optimizing disease management, reducing veterinary expenses, and promoting resource-efficient agricultural practices. As we embrace this intersection of technology and veterinary science, the ripple effects encompass not only the well-being of bovine populations but also the foundation of ecologically-conscious and economically-viable farming approaches. Recent advances in Artificial Intelligence have not only demonstrated their potential in various medical applications but have also paved the way for innovative approaches in disease detection, including predictive systems for general medicine (Nadeem et al., 2023). Binary classification techniques have emerged as the linchpin for distinguishing crucial attributes from complex data, facilitating the accomplishment of precise objectives. Within this context, an array of classification models, such as the Random Forest (RF) (Dutta et al., 2022), Extreme Gradient Boost (XGB) (Gertz et al., 2020), Logistic Regression (LR) (Becker et al., 2020), and Single Perceptron Model (SPM) (Bauer et al., 2022), has gained prominence among researchers in the same domain. To ensure the optimal performance of each machine learning model, rigorous hyper-parameter tuning was performed (Nadeem et al., 2023). The performance evaluation of the models was based on multiple accuracy factors, including accuracy, precision, F1-score, recall, and the area under the curve (AUC-ROC). The results of this analysis highlight the superiority of the random forest model, which outperformed the other models across all accuracy metrics for detecting and classifying bovine diseases and events.
This research paper offers significant contributions to the field of agricultural informatics and bovine health management. By harnessing the potential of machine learning and artificial intelligence, we present an innovative approach to improve disease detection accuracy, reduce economic losses, and enhance overall dairy productivity. The successful implementation of the proposed automated disease detection system has the potential to revolutionize the dairy industry, benefiting farmers, veterinarians, and stakeholders by promoting early intervention, improving animal welfare, and maximizing dairy production. ML-based systems empower farmers with data-driven insights and timely interventions, ultimately leading to improved animal welfare, enhanced productivity, and reduced economic losses in the dairy industry.
The remaining paper is structured as follows. The related study in the field of bovine welfare events detection system is discussed in Section Related Work. Then, methods and the implementation of ML models. Next the data sets utilized for the training and testing phases of models are discussed. After that the performance analysis and a comparison of AI model evaluations are highlighted. At last, we draw the Conclusions and discussed the future work.
Related work
Machine learning algorithms have been investigated by a number of researchers to identify or forecast the start of various health conditions. Each study, however, has concentrated on just one to three illnesses.
The author (Wagner et al., 2020) investigates the application of machine learning algorithms to detect abnormal behavior in dairy cows experiencing Sub-Acute Ruminal Acidosis (SARA). The authors point out that although new sensor technology makes it possible to continuously monitor animal behavior, the transition to abnormal animal activity is still challenging to spot. In order to identify small behavioral changes that may occur just before clinical manifestations of an illness, the author suggests using machine learning algorithms. Number of machine learning techniques evaluated, including K Nearest Neighbors for Regression (KNNR), Decision Tree for Regression (DTR), Multi-Layer Perceptron (MLP), Long Short-Term Memory (LSTM), and a technique that relies on the assumption that activity is consistent from day to day. The best performance was discovered by KNNR, which detected 83% of SARA cases (true-positives), but it also generated 66% of false-positives, which restricts its practical application. The study (Wagner et al., 2020) finding shows that machine learning can be used to identify behavioral anomalies and that more advancements might likely be made by using machine learning on extremely huge data sets at the animal level as opposed to the group level.
Similarly, the use of machine learning algorithms for early prediction of common disorders as digestive disorder (Zhou et al., 2022), includes lameness, mastitis, and metritis in dairy cows monitored by automatic systems for 280 cows where 131 were the sick cows. The authors developed a predictive model based on eight machine learning algorithms, including XGB, random forest, KNN and R part algorithms and evaluated the performance of the model using various metrics, such as precision, accuracy, and area under the curve. The results showed that all models achieved high levels of accuracy in predicting common disorders in dairy cows, the accuracy of eight algorithms in range of 65% to 84%. However the R part best predicts the disorders with an accuracy, precision, and AUC of 81.58%, 92.86%, and 0.908, respectively. The use of machine learning algorithms in dairy farming can help farmers identify and treat common disorders early, leading to improved cow health and productivity. This paper (Zhou et al., 2022) highlights the importance of using advanced technologies such as machine learning algorithms for improving dairy farming practices and ensuring the health and well-being of dairy cows.
In the study (Lardy et al., 2023), machine learning techniques were harnessed to distinguish between pathological, reproductive, and stress conditions in cows using sensor-based activity data. The research aimed to develop accurate models for condition identification based on cow activity patterns. Various machine learning algorithms, including Random Forest, Support Vector Machine, and Neural Network, were employed to create predictive models. The data-set encompassed diverse behavioral attributes and physiological measurements includes five data-sets (120,000 cow days) from experimental or commercial farms were observed, with 28 to 300 Holstein cows per data-set and observations lasting from 1 to 12 months. Sensors captured the per-hour duration of key cow activities, and the distribution of the activity level was described by 21 time-domain or frequency-domain features. The models were trained and evaluated on a range of accuracy metrics, including precision, recall, F1-score, and AUC-ROC. Results highlighted the potential of machine learning models in precisely discerning different conditions. Notably, the Random Forest model exhibited promising performance, achieving an average accuracy of 87%. The study (Lardy et al., 2023) underscores the feasibility of using machine learning for effective classification of conditions in dairy herd management.
Materials and Methods
Data set uses in this paper have prior target classes so supervised machine learning is preferred for ML models, the targeted class is either be 0 to accrue the state of not conduction of individual event/disease, or would be the 1 to represent the presence of particular event or disease for that time instance so the classification of binary nature is observed. Figure 1 depicts the overall flow of proposed system, at the initial stage, data from four distinct data-sets is acquired for input. Following this, the data undergoes a preprocessing phase before progressing to the allocation of data-sets for training and testing, maintaining a ratio of 80% for training and 20% for testing. This partitioning of data-sets serves as the foundation for subsequent phases, wherein the data-sets are harnessed to train a variety of Machine Learning models. Through Python 3.10.6, and utilizing libraries including SciKit-Learn v1.1.3, numpy v1.23.5, pandas v1.2.4 and tensor flow v2.5, the models are systematically trained. Post-training, a rigorous validation process ensues to comprehensively evaluate the performance of these models. This evaluation culminates in a comparative analysis of the models, conducted across a spectrum of accuracy parameters. The orchestration of this entire ML process embodies a meticulously structured approach, one that capitalizes on cutting-edge tools and methodologies to extract meaningful insights from complex data-sets.
Data acquisition
Data gathered from four sets of univariate time series containing 386 different Holstein cattle. Data-sets 1 and 2 were obtained from the French experimental farm INRAE Herbipôle in Marcenat. Data from 28, 28, 30, and 300 cows, which were observed for six months, two months, forty days, and one year, respectively, are included in Data-sets 3 and 4, which are commercial farms in Europe (Lardy et al., 2023).
Data preprocessing
The data preprocessing is a vital role on model, it determines the ultimate performance of machine learning models (Habiband Khursheed, 2022), which includes data cleaning, data normalization, data visualization, balancing of data is required as each event/disease of an individual data-set have the unbalanced numbers of the normal and occurrence state of the events as shown in Table 1, and at last it includes the attribute selection as the data-set is composed of 19 different labels where the most 3 prominent attribute which correlated to each single event and disease are chosen, label as EAT, REST and IN-ALLEYS.
Table 1: Unbalanced distribution of events in different data-sets.
|
Events |
Dataset-1 |
Dataset-2 |
Dataset-3 |
Dataset-4 |
||||
|
0s |
1s |
0s |
01s |
0s |
1s |
0s |
1s |
|
|
Estrus |
106681 |
40079 |
40079 |
40079 |
168 |
168 |
25601 |
624 |
|
Mastitis |
107449 |
40175 |
40175 |
40175 |
72 |
72 |
0 |
0 |
|
Lameness |
107569 |
39863 |
39863 |
39863 |
384 |
384 |
0 |
0 |
|
Calving |
107473 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
|
Acidosis |
0 |
33743 |
33743 |
33743 |
6504 |
6504 |
0 |
0 |
Training and testing set
The training and testing phase stand as a pivotal pillar within our research framework, serving as the bedrock for crafting and rigorously evaluating specialized machine learning models aimed at precision-driven bovine disease and event detection. The division of our data-set into distinct training and testing subsets, using a meticulous 80-20 ratio allocation, ensured a robust foundation for model development and assessment. Notably, this approach accommodated the amalgamation of four diverse data-sets, each contributing to a holistic representation of real-world scenarios. Amidst this intricate landscape, a carefully curated ensemble of machine learning models, encompassing random forest, extreme gradient boost, linear regression, and single perceptron, were meticulously fine-tuned and calibrated within the training phase. This strategic process harnessed the vast dimensions of the combined data-sets, thus amplifying the models’ learning potential.
ML models
ML algorithms have been widely used in the diagnosis, prediction, and other fields of Bovine due to their powerful complex data processing capabilities (Liu et al., 2023). The choice of classifier models is constrained to those that are well-known and commonly employed, 4 ML models were deployed to identify the best model for detection of event/disease based on data.
Random forest
A versatile machine learning technique called random forest ensures accuracy and stability during training by building several decision trees. It corrects for overfitting and can handle large datasets without variable deletion. Its ease of use and flexibility make it a preferred choice for predictive modelling challenges. The SciKit-Learn package The RF classifier originates from the Random forest classifier, and the grid search CV method is used to hyper-parameter tune the three key parameters with a cross validation (CV) of 20, n-estimators with a range between 10 and 1500, and max-depth in a range between 1 and 30. The maximum depth is 7, the n-estimator value is 550, and entropy is the determining factor. Entropy, as shown in Equation 1, describes how chaotic or unpredictable a given split is. The default settings apply to every other parameter.
Here Pj is the probability of class J.
Extreme gradient boost
XGBoost is a powerful machine learning algorithm that utilizes gradient boosting and decision trees as weak learners in an ensemble framework. Its performance, efficiency, and scalability make it a popular choice for large-scale data handling. Its optimization strategies, including gradient-based learning and tree pruning, contribute to its accuracy in predictive modeling tasks. The SciKit-Learn package xgboost is used to construct the XGB classifier, the algorithm mathematically represented in Equation 2 by the second order Taylor expansion equation (Jing et al., 2022). XGB model with hyper-parameter of n-threads parallel processes in number (n-threads) = 1, objective linear logistic, with the method of GridSearchCV is employed for the hyper-parameter tuning with CV of 20, max-depth is range of 5 to 25, n-estimators are in range of 100 to 500, eta ranges from 0.01 to 0.7 where the optimist value for max-depth is 10, n-estimators of 455 and eta 0.46 remaining all other parameters are the default values.
Here in Equation 2, yt is the prediction for the p-th feature example at the t-th increment. An objective loss function L is used to express the mathematical equation.
Logistic regression
Logistic regression is a statistical method used for binary outcomes classification, estimating probabilities using a logistic function on linear input features. It’s effective when data is linearly separable and target variable is dichotomous, providing a robust framework for classification and interpretability in academic and applied settings. The SciKit-Learn package logistic regression is used, mathematically shown in Equation 3. The method of Grid Search CV is employed for the hyper-parameter tuning with CV of 20, the regularization penalty is offered of (none, l1, l2), similarly the C value which is a penalty term, meant to disincentives and regulate over-fitting, is in range of (100, 10, 1.0, 0.01, 0.001), solvers are (lbfgs, liblinear, sag and saga) and max-iter of (1 to 1000), where the optimist value for regularization penalty is l2, value for the C is 0.001, solver is saga and max-iter of 554 are observed, the all other remaining parameters are set as default values.
Here h is the output function of logistic regression and R is the hypothesis which is WX + B, where W is the slope, X is he independent variable and B is the y-intercept accordingly.
Perceptron
A key idea in machine learning is the perceptron model, a neural network that predicts binary classifications using a feature vector and weights. Despite its simplicity, this model served as a foundational idea in the field of machine learning and helped pave the way for more intricate neural networks. A simple perceptron model by SciKit-Learn package perceptron is used as shown in equation 4, trained using the stochastic gradient descent optimization algorithm and activation function by default is step function with an eta of 0.0001 and initially by default weights are uniform and other remaining parameters are default values.
Z is the current node’s output, f is the activation function, xi is the input from the node in the preceding layer with weight wi, and b is the bias to adjust the spectrum of the summation value.
Evaluation
How well a machine learning model responds to new, and unexplored data is what determines how effective it is. An evaluation is the last action. The F1-score, Precession, and Recall values were examined along with the accuracy scores to find the ideal configuration strategy for machine learning models (Habib and Khursheed, 2022). In order to further confirm the results, the receiver operating characteristic (ROC) and area under the curve (AUC) are also displayed against true positive and false negative data.
Dataset description
Several data-sets for Bovine health and disease detection are available, the data-set used for this study is collected from the Cow-View system (GEA Farm Technology, Bönen, Germany), Data is composed of four data-sets the data-sets are structured as follows: 386 Holstein breed unique cows with a tag id of 3 to 5 digits, date, hourly scale aggregation of individual activity, time spent in Eating, Resting and in walking ‘In Alleys’ are in unit seconds for each hour scale. Finally, for each of the 11 types of events, a Boolean is provided, i.e., 1 indicates that this kind of incident was reported for this hour, whereas 0 indicates that it was not. Six of these 11 types of events lameness, mastitis, LPS (administration of lipopolysaccharide in the mammary gland, an experimental treatment to induce udder inflammation), sub-acute ruminal acidosis, other diseases (such as colic, diarrhea, ketosis, milk fever, or other infectious diseases), and accidents (such as retained placenta or vaginal laceration) were linked to health and were divided into three states. Estrus and calving were considered to be two reproductive events. Animal mingling, disruption (i.e., minor interventions on animals like late feeding, alarm tests, and fill beds), and marginal management modifications (ration alterations, fill beds) were three occurrences that caused stress. In addition, a Boolean sum up whether this hour was considered as normal or not (Lardy et al., 2022).
Data-sets 1 and 2 are from INRAE Herbipôle experimental farm, Marcenat, France data-sets 3 and 4 are European commercial farms, contains data on respectively 28, 28, 30 and 300 cows monitored for 6 months, 2 months, 40 days and one year with the approximately losses of 12.45% with the data for 107665 hours, 0.2% withholding data for 40246 hours, 10.4% for 26224 hours and 16.47% with the data for 2177207 hours (Lardy et al., 2022). The frequency of events observed in each data-sets is in Table 2.
Table 2: Frequency of events occurrence in data-sets.
|
Events |
Occurrence |
|||
|
Dataset-1 |
Dataset-2 |
Dataset-3 |
Dataset-4 |
|
|
Accidental events |
0 |
0 |
0 |
15 |
|
Calving |
8 |
0 |
0 |
171 |
|
Estrus |
41 |
7 |
26 |
257 |
|
Lameness |
4 |
16 |
0 |
114 |
|
LPS injection |
27 |
0 |
0 |
0 |
|
Management changes |
0 |
168 |
0 |
2581 |
|
Mastitis |
9 |
3 |
0 |
32 |
|
Mixing |
72 |
0 |
0 |
0 |
|
Other disturbance |
173 |
671 |
0 |
12223 |
|
Other disease |
10 |
8 |
0 |
66 |
|
Ruminal acidosis |
0 |
271 |
0 |
0 |
The events covered for the training and testing on ML models are: Calving, Estrus, Mastitis, and Lameness, other remaining factors are for the farm’s observed during the data gathering.
Results and Discussion
The ideal setup strategy for machine learning models is decided using accuracy scores and other metric variables. Accuracy, recall, precision, F1 score, and AUC are the variables utilized to evaluate the efficacy of our trained models, as shown in Equations 5, 6, 7, and 8 accordingly. Since ROC is a stronger performance evaluator than a confusion matrix, it was used in terms of AUC for our performance evaluation of several machine learning techniques. The true positive and false positive values in the ROC graph are used to calculate the AUC accuracy, which can provide accurate accuracy numbers despite differences in classifier performance. The mentioned metrics, which are measured in this work as (FN), (FP), (TN), and (TP), can be used to assess the effectiveness of the classifiers.

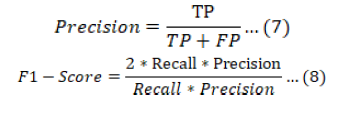
Where TP stands for true positives, or the number individuals who were accurately projected to be positive, and TN stands for true negatives, or the number of the individuals who were accurately expected to be negative; False positives (FP) are the number of individuals who were mistakenly forecasted to be positive but were actually negative, while false negatives (FN) are the number of individuals who were mistakenly predicted to be negative but were actually positive (Liu et al., 2023).
Data-set 1
Figures 2, 3, 4, 5 represents the graphical results of event Estrus, Calving, Mastitis, Lameness covered for the training and testing on different ML models for the data-set 1, respectively.
Data-set 2
Figures 6, 7, 8 and 9 represents the graphical results of event Estrus, Acidosis, Mastitis, Lameness covered for the training and testing on different ML models for the data-set 2, respectively.
Data-set 3
Events covered for the training and testing on ML models for this data-set is estrus only similarly the results are accordingly. Figure 10 represents the graphical results of the event Estrus for different ML models in the data-set 3.
Data-set 4
Figures 11, 12, 13, 14 represents the graphical results of event Estrus, Calving, Mastitis, Lameness covered for the training and testing on different ML models for the data-set 2, respectively.
The Figure 15 encapsulates the comprehensive performance evaluation of model across distinct events detection scenarios. The Random Forest Classifier (RFC) exhibited robust performance in event classification as compare to other models, demonstrating high accuracy, precision, recall, F1 score, and AUC metrics across various data-sets. For Estrus detection, the RFC model showcased consistent excellence, with an average accuracy of 91.60% and a balanced F1 score of 94.50%. Calving prediction achieved exceptional results with an average accuracy of 95.89% and an impressive F1 score of 99.50%. Mastitis identification demonstrated high accuracy, averaging at 92.96%, and achieved remarkable recall at 94.67%. Lameness detection exhibited consistent performance across data-sets, with an average accuracy of 91.47% and an F1 score of 98.67%. Acidosis prediction yielded promising results with an average accuracy of 79.88%, showing potential in early event detection. These outcomes collectively underscore the effectiveness of the RFC model in event detection, across diverse contexts and data-sets, making it a robust choice for real-time monitoring and prediction.
Conclusions and Recommendations
In this paper, a comprehensive analysis of ML models for early bovine disease and event detection is presented, illuminating their pivotal role in advancing global dairy farming. Our findings underscore the trans-formative potential of ML-driven insights, empowering farmers with timely interventions, heightened productivity, and reduced economic losses in the dairy industry.
This research underscores the effectiveness of ML models in early disease detection and precise classification, emphasizing the urgency of timely identification for animal health, economic sustainability, and a thriving dairy sector. The integration of ML in bovine disease detection marks a substantial stride toward data-driven agricultural practices, reshaping efficiency, productivity, and animal welfare. We employed four prominent ML models – Random Forest, Perceptron Model, Extreme Gradient Boost, and Linear Regression – crafting disease detection systems from a comprehensive data-set amalgamating historical health records and event timestamps. Significantly, the Random Forest model emerges as a standout performer, consistently excelling across critical accuracy metrics – f1 score, accuracy with the value of 98.25 in detecting Mastitis, precision with the value of 97 in mastitis, recall with the score of 100, and AUC with 0.999 – in various individual data-sets. This research echoes the promise of data-centric innovation, heralding a future where technology seamlessly intertwines with agricultural tradition to forge a resilient, efficient, and compassionate dairy farming landscape. As we continue to harness the power of data in agriculture, we can look forward to a future where dairy farming is not only productive and profitable but also ethical and environmentally friendly.
Acknowledgement
Thanks and gratitude to all authors
Novelty Statement
The study demonstrated the impact of different ML models to cope with the earlier detection of such events and disease to cure the essentials in the time. The results shows that the ML model Random Forest out perform as compare to other models and these extracts will certainly be used in future applications regarding disease and events detection.
Author’s Contribution
Ghalib Nadeem: Formal analysis and writing original draft preparation, carried out planning of study, experimentation, and analysis.
Muhammad Irfan Anis: Supervised, helped in carried out planning of study and revision.
Conflict of interest
The authors have declared no conflict of interest.
References
Alexandra, C. and Green, A. C., Lidfors, L. M., Lomax, S., Favaro, L., & Clark, C. E. (2021). Vocal production in postpartum dairy cows: Temporal organization and association with maternal and stress behaviors. J. Dairy Sci., 104(1): 826-838. www.sciencedirect.com /science/article/pii/ S0022030220308778.
Ali, S.K., Catal, C., Kaya, A., and Tekinerdogan, B.. 2020. Development of a recurrent neural networks-based calving prediction model using activity and behavioral data. Comp. Electron. Agric., www.sciencedirect.com /science/article/abs/pii /S0168169919312220.
Alipio, M. and M.L. Villena. 2022. Intelligent wearable devices and biosensors for monitoring cattle health conditions: A review and classification. Smart Health, pp. 100369. https://doi.org/10.1016/j.smhl.2022.100369
Arcidiacono, C., Massimo Mancino, and S. M. C. Porto. 2020. Moving mean-based algorithm for dairy cow’s oestrus detection from uniaxial-accelerometer data acquired in a free-stall barn. Comp. Electron. Agric., 175: 105498. https://doi.org/10.1016/j.compag.2020.105498
Bauer, E.A. and Jagusiak, W., 2022. The use of multilayer perceptron artificial neural networks to detect dairy cows at risk of ketosis. Animals, 12(3): 332. https://doi.org/10.3390/ani12030332
Becker, C.A., Aghalari, A., Marufuzzaman, M., & Stone, A. E. 2020. Predicting dairy cattle heat stress using machine learning techniques. J. Dairy Sci. Elsevier, 104(1), 501-524. https://www.sciencedirect.com /science/article/pii/S0022030220308663.
Cantor, M.C., H.M. Goetz, K. Beattie and D.L. Renaud. 2022. Evaluation of an infrared thermography camera for measuring body temperature in dairy calves. JDS Commun., 3(5): 357-361. https://doi.org/10.3168/jdsc.2022-0227
Das, S., A. Shaji, D. Nain, S. Singha, M. Karunakaran and R.K. Baithalu. 2023. Precision technologies for the management of reproduction in dairy cows. Trop. Anim. Health Prod., 55(5): 286. https://doi.org/10.1007/s11250-023-03704-2
Dutta, D., D. Natta, S. Mandal and N. Ghosh. 2022. Monitor: An IoT based multi-sensory intelligent device for cattle activity monitoring. Sensors and actuators A: Physical, 333: 113271. https://doi.org/10.1016/j.sna.2021.113271
García, R., J. Aguilar, M. Toro, A. Pinto and P. Rodríguez. 2020. A systematic literature review on the use of machine learning in precision livestock farming. Comp. Electron. Agric., 179: 105826. https://doi.org/10.1016/j.compag.2020.105826
Gertz, M., Grobe-Butenuth, K., Junge, W., Maassen-Francke, B., Renner, C., Sparenberg, H., and Krieter, J. . 2020. Using the XG boost algorithm to classify neck and leg activity sensor data using on- farm health recordings for locomotor-associated diseases, computers and electronics in agriculture- X-MOL.” Science Direct, Computer and Electronics. https://doi.org/10.1016/j.compag.2020.105404
Ghafoor, N.A. and B. Sitkowska. 2021. MasPA: A machine learning application to predict risk of mastitis in cattle from AMS sensor data. Agri Engineering, 3(3): 575–584. https://doi.org/10.3390/agriengineering3030037
Habib, B. and F. Khursheed. 2022. Performance evaluation of machine learning models for distributed denial of service attack detection using improved feature selection and hyper-parameter optimization techniques. Concurr. Comput. Pract. Exp., 34(26): e7299. https://doi.org/10.1002/cpe.7299
Haladjian, J., J. Haug, S. Nüske and B. Bruegge. 2018. A wearable sensor system for lameness detection in dairy cattle. Multimod. Technol. Interact., 2(2): 27. https://doi.org/10.3390/mti2020027
Han, C.S., U. Kaur, H. Bai, B.R. Dos Reis, R. White, R.A. Nawrocki and S. Priya. 2022. Invited review: Sensor technologies for real-time monitoring of the rumen environment. J. Dairy Sci., 105(8): 6379-6404. https://doi.org/10.3168/jds.2021-20576
Heirbaut, S., D.B. Jensen, X.P. Jing, B. Stefańska, P. Lutakome, L. Vandaele and V. Fievez. 2022. Different reticuloruminal pH metrics of high-yielding dairy cattle during the transition period in relation to metabolic health, activity, and feed intake. J. Dairy Sci., 105(8): 6880-6894. https://doi.org/10.3168/jds.2021-21751
Huang, S.Z., Y.S. Chen, J.T. Hsu and T.T. Lin. 2023. Dairy cow health status evaluation based on multi-sensor data fusion and machine learning. In 2023 ASABE Annual International Meeting (p. 1). Am. Soc. Agric. Biol. Eng., https://doi.org/10.13031/aim.202300293
Hyodo, R., T. Nakano and T. Ogawa. 2023. Deep multi-stream network for video-based calving sign detection.
Jing, X., Q. Zou, J. Yan, Y. Dong and B. Li. 2022. Remote sensing monitoring of winter wheat stripe rust based on mRMR-XGBoost algorithm. Remote Sens., 14(3): 756. https://doi.org/10.3390/rs14030756
Kang, X., S. Li, Q. Li and G. Liu. 2022. Dimension-reduced spatiotemporal network for lameness detection in dairy cows. Comp. Electron. Agric., 197: 106922. https://doi.org/10.1016/j.compag.2022.106922
Lardy, R., Q. Ruin and I. Veissier. 2023. Discriminating pathological, reproductive or stress conditions in cows using machine learning on sensor-based activity data. Comp. Electron. Agric., 204(2023): 107556. https://doi.org/10.1016/j.compag.2022.107556
Lardy, R.M.M., N. Mialon, Y. Wagner, Gaudron, B. Meunier, K.H. Sloth, D. Ledoux, Veissier, I. 2022. Understanding anomalies in animal behaviour: Data on cow activity in relation to health and welfare. Anim. Open Space, 1(1): 100004. https://doi.org/10.1016/j.anopes.2022.100004
Leliveld, L., C. Brandolese, M. Grotto, A. Marinucci, N. Fossati, D. Lovarelli and G. Provolo. 2021. Integrating multi-sensor information for the real-time automatic monitoring of barn environment and dairy cattle behaviour. Available at SSRN 4511074.
Liu, R., Z. Xu, J. Teng, X. Pan, Q. Lin, X. Cai, S. Diao, X. Feng, X. Yuan, J. Li and Z. Zhang. 2023. Evaluation of six machine learning classification algorithms in pig breed identification using SNPs array data. Anim. Genet., 54(2): 113-122. https://doi.org/10.1111/age.13279
Matera, R., G. Di Vuolo, A. Cotticelli, A. Salzano, G. Neglia, R. Cimmino and S. Biffani. 2022. Relationship among milk conductivity, production traits, and somatic cell score in the Italian Mediterranean buffalo. Animals, 12(17): 2225. https://doi.org/10.3390/ani12172225
Nadeem, G., Y. Rehman, A. Khaliq, H. Khalid and M.I. Anis. 2023. Artificial intelligence based prediction system for general medicine. 4th International Conference on Computing, Mathematics and Engineering Technologies (iCoMET), Sukkur, Pakistan, pp. 1-6. https://doi.org/10.1109/iCoMET57998.2023.10099078
O’Leary, N.W., Byrne, D.T., O’Connor, A.H., and Shalloo, L. 2020. Invited review: Cattle lameness detection with accelerometers. J. Dairy Sci., 103(5): 3895–3911. https://doi.org/10.3168/jds.2019-17123
Post, C., C. Rietz, W. Büscher and U. Müller. 2020. Using sensor data to detect lameness and mastitis treatment events in dairy cows: A comparison of classification models. Sensors, 20(14): 3863. https://doi.org/10.3390/s20143863
Riaboff, L., L. Shalloo, A.F. Smeaton, S. Couvreur, A. Madouasse and M.T. Keane. 2022. Predicting livestock behaviour using accelerometers: A systematic review of processing techniques for ruminant behaviour prediction from raw accelerometer data. Comp. Electron. Agric., 192: 106610. https://doi.org/10.1016/j.compag.2021.106610
Roche, S.M., Ross, J. A., Schatz, C., Beaugrand, K., Zuidhof, S., Ralston, B., and Olson, M. 2023. Impact of dystocia on milk production, somatic cell count, reproduction and culling in Holstein dairy cows. Animals, 13(3): 346. https://doi.org/10.3390/ani13030346
Santos, C.A., N.M.D. Landim, H.X. de Araújo and T.P. Paim. 2022. Automated systems for estrous and calving detection in dairy cattle. Agri Eng., 4(2): 475–482. https://doi.org/10.3390/agriengineering4020031
Themistokleous, K.S., I. Papadopoulos, N. Panousis, A. Zdragas, G. Arsenos and E. Kiossis. 2023. Udder ultrasonography of dairy cows: Investigating the relationship between echotexture, blood flow, somatic cell count and milk yield during dry period and lactation. Animals, 13(11): 1779. https://doi.org/10.3390/ani13111779
Van Nuffel, A., I. Zwertvaegher, L. Pluym, S. Van Weyenberg, V.M. Thorup, M. Pastell, B. Sonck and W. Saeys. 2015. Lameness detection in dairy cows: Part 1. How to distinguish between non-lame and lame cows based on differences in locomotion or behavior. Animals An Open Access J. MDPI, 5(3): 838–860. https://doi.org/10.3390/ani5030387
Wagner, N., V. Antoine, M.M. Mialon, R. Lardy, M. Silberberg, J. Koko and I. Veissier. 2020. Machine learning to detect behavioural anomalies in dairy cows under subacute ruminal acidosis. Comp. Electron. Agric. 170: 105233. https://doi.org/10.1016/j.compag.2020.105233
Wang, J., M. Bell, X. Liu and G. Liu. 2020. Machine-learning techniques can enhance dairy cow estrus detection using location and acceleration data. Animals (Basel), 10(7): 1160. https://doi.org/10.3390/ani10071160
Wang, Y., S. Li, H. Zhang and T. Liu. 2022. A lightweight CNN-based model for early warning in sow oestrus sound monitoring. Ecol. Inf., 72: 101863. https://doi.org/10.1016/j.ecoinf.2022.101863
Zheng, Z., Zhang, X., Qin, L., Yue, S., and Zeng, P.. 2023. Cows’ legs tracking and lameness detection in dairy cattle using video analysis and Siamese neural networks. Comp. Electron. Agric., 205: 107618. https://doi.org/10.1016/j.compag.2023.107618
Zhou, X., Xu, C., Wang, H., Xu, W., Zhao, Z., Chen, M., and Huang, B. 2022. The early prediction of common disorders in dairy cows monitored by automatic systems with machine learning algorithms. Animals, 12(10): 1251. https://doi.org/10.3390/ani12101251
To share on other social networks, click on any share button. What are these?