Advances in Animal and Veterinary Sciences
Research Article
Relationship between Phenotypic and Genotypic Antimicrobial Resistance of Escherichia coli Isolates from Mastitic Milk
Samah El-sayed M, Soliman M Soliman*, Adel Abdel-Azim Fayed, Samia Abd El-hamid Ahmed
Department of Medicine and Infectious Diseases, Faculty of Veterinary Medicine, Cairo University, Egypt.
Abstract | Bovine mastitis is the predominant problem in dairy farms worldwide which caused mainly by Escherichia coli (E. coli) as one of the main causes of what is called “environmental mastitis”. A total of 68 E. coli isolates from 205 raw milk samples of Holstein cows with mastitis in different dairy farms from different governorates by bacteriological isolation and 63 by PCR were investigated for the E. coli 16S rRNA and rfbEO157 encoding gene as Shiga toxin-producing E. coli (STEC). The occurrence of E. coli O157 in mastitic cows was 3% within E. coli isolates. Molecular investigation of extended-spectrum β-lactamases (ESBLs) and plasmid-mediated AmpC β-lactamases (PABLs) encoding genes reported in all of the isolates (100%) encoded TEM-type ESBLs, none of which (0%) encoded OXA-type ESBLs, on the other hand, CTX-M-type ESBLs and SHV-type β-lactamases were encoded in 34/63 (53.9%) and 3/63 (4.7%) of the ESBL isolates, respectively and 27% exhibited CMYІІ-type PABLs. Plasmid-mediated colistin resistance encoding gene (mcr-1) was expressed in 1.6% of E. coli isolates. All E. coli isolates exhibited antibiotic multi-resistances with higher resistance to tetracycline and Trimethoprim-Sulfamethoxazole (45.7% and 37.3%, respectively), while the lowest resistance was observed for Amoxicillin/clavulanate (10.1%). Phenotypic resistance to extended-spectrum cephalosporins (ESCs) revealed that 42.3% of these strains were resistant to (cefotaxime and cefquinome), 15% resistant to Cefoxitin, while 32.2% were resistant to ceftazidime. Conclusively, E. coli was found to be the major cause of bovine mastitis treatment failure due to the multidrug resistance to most newly developed cephalosporins (third and fourth generations).
Keywords | Bovine mastitis, Milk, Antimicrobial Resistance, ESBLs, PABLs and mcr-1.
Received | March 30, 2021; Accepted | April 10, 2021; Published | July 01, 2021
*Correspondence | Soliman M Soliman, Department of Medicine and Infectious Diseases, Faculty of Veterinary Medicine, Cairo University, Egypt; Email: soliman450@yahoo.com
Citation | M El-Sayed S, Soliman SM, Fayed AAA, Ahmed SAH (2021). Relationship between phenotypic and genotypic antimicrobial resistance of escherichia coli isolates from mastitic milk. Adv. Anim. Vet. Sci. 9(8): 1223-1232.
DOI | http://dx.doi.org/10.17582/journal.aavs/2021/9.8.1223.1232
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2021 Soliman et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRoDUCTION
Mastitis is a major concern for dairy producers causing significant economic losses for the dairy industry. The most frequently isolated causative agent related to bovine intramammary infection is Escherichia coli (E. coli) (Keane et al., 2013; Olde et al., 2008; Bradley et al., 2007), Because E. coli is a widespread environmental pathogen, can invade the udder. The decrease in the incidence of clinical mastitis has a positive impact on animal health, animal welfare, antimicrobial usage, work pleasure, and net farm return (Trevisi et al., 2014). Herd management factors, such as milking technique and hygiene standards, were associated with variations in distributions of a mastitis-causing pathogen in the herd (Barkema et al., 1999; Dufour et al., 2011; Piepers et al., 2011; Levison et al., 2016).
Clinical mastitis can be caused mainly by coliform infections (De Vliegher et al., 2012). A wide range of systemic disease severity, from mild to severe with systemic signs including dehydration, shock, and even death (Wenz et al., 2001). As well as zoonotic public health impact on human especially Shiga-toxigenic E. coli (STEC) strains including O157 causing bovine mastitis (Lin et al., 2011).
Many problems facing the dairy industry requires antimicrobial therapy, Mastitis is one of them (Grave et al., 1999). Which is usually employed in treating/preventing mastitis, such as β-lactams, sulphonamides, quinolones, macrolides and tetracyclines (Bengtsson et al., 2009; Mathew et al., 2007; McEwen and Fedorka-Cray, 2002).
The misuse of antibiotics caused drug resistance and treatment failures in many cases, especially for multidrug-resistant bacteria (Suojala et al., 2013; Sweeney et al., 2018). Carattoli (2008) has announced the antimicrobial resistant E. coli strains increase within animals and claimed these animals to be a reservoir of such strains for humans and the environment. Potential transmission of resistant E. coli within animals and humans can occur through various pathways, as the food chain (Poirel et al., 2018).
Extended-spectrum β-lactamases (ESBLs) producing E. coli, which shows resistance to penicillins, aminopenicillins, and cephalosporins, including the third (ceftiofur) and fourth (cefquinome) generations, has been commonly isolated from food-producing animals with global veterinary and public health issues (Seiffert et al., 2013; Poirel et al., 2018). ESBLs that inactivates ESCs were graded as class A (TEM, SHV and CTXM) and class D (OXA) β-lactamases, While plasmid AmpC β-lactamases (PABLs) belonged to class C (CMYII) confer resistance to a wide variety of β-lactams, primarily 7-a-methoxy-cephalosporins (Cephamycins) such as cefoxitin (Livermore and Woodford, 2006; Jacoby, 2009). Antibiotics used for humans and animals are closely related, abuse of these drugs resulted in the development of multidrug-resistant bacteria (Cantas et al., 2013; Walther et al., 2017). So, for efficient control and treatment of mastitis; the causative agents of IMI in dairy herds need to be well-identified. Antimicrobial susceptibility determined in vitro has been considered as a pre-requisite for treatment. However, in vitro activity does not guarantee in vivo effectiveness in bovine mastitis treatment (Pyörälä, 2009).
Colistin, a member of polymyxins (polymyxin E), is the main drug for E. coli (Kempf et al., 2013; Poirel et al., 2017). But, Colistin resistance was identified due to the emergence of highly transmissible plasmid-mediated colistin-resistant (mcr-1) gene in E. coli strains obtained from animals, food, and patients from China (Liu et al., 2016). This resistance has created global issues due to the high transmission rate of the mcr-1 gene to epidemic strains of Enterobacteriaceae and thus hinders the effectiveness of colistin in humans (Rebelo et al., 2018).
The objectives of this study were to identify the impact of multidrug resistance development of E. coli strains isolated from mastitic dairy cow’s milk, evaluate phenotypic antibiotic resistance profile of isolated strains and their association to genotypic antimicrobial resistance to provide efficient treatment.
MATERIALS AND METHODS
Sample Collection
205 pooled milk samples were collected using the California mastitis test (CMT) from 205 mastitic dairy cows from five dairy farms located in Fayoum, Ismailia, El-sharkia, Alexandria and Giza governorates between November 2019 and October 2020. Milk samples (approximately 15 ml) were aseptically drawn from each cow immediately according to the National Mastitis Council, 1990 then samples were transferred to the laboratory for further examination.
Phenotypic Identification
Milk samples were cultured in Eosin Methylene Blue agar media (EMB) (Oxoid). Agar plates were incubated at 37ºC, and the bacterial growth was evaluated after 24 and 48 hrs. Using phenotypic differentiation of bacterial species presumptively based on colony morphology and Gram’s staining (David, 2011).
Genotypic Identification
The genomic DNA of all E. coli strains was extracted (Kang et al., 2004) and stored at -20°C for detection of genes encoding for 16srRNA, rfbEO157 encoding virulence gene and antibiotic resistance genes of E. coli strains isolated from mastitic milk samples (Table 1).
PCR amplification of 16srRNA encoding gene was performed according to Wang et al. (2002) as illustrated in Table 1. The reaction was performed in a volume of 25 μl containing 12.5 μl of 2X Qiagen Multiplex PCR Master Mix (Qiagen GmbH, Hilden, Germany), 0.5 µl (10pmol/µl) concentrations of each primer, and 3 μl of DNA template. The amplified PCR products were subjected to electrophoresis using 1.5% agarose gel.
Table 1: Oligonucleotide primers used for conventional PCR assay.
| Target gene | Primer sequence | Amplicon size (bps) | Source |
|
Detection of E. coli isolates |
|||
| 16S rRNA |
CCCCCTGGACGAAGACTGAC ACCGCTGGCAACAAAGGATA |
401 |
Wang et al. (2002) |
|
β- lactamase genes |
|||
|
O157 (rfbEo157) |
CGG ACA TCC ATG TGA TAT GG TTG CCT ATG TAC AGC TAA TCC |
259 |
Possé et al. (2007) |
|
E. coli O157 gene |
|||
|
bla SHV
|
CTT TAT CGG CCC TCA CTC AA AGG TGC TCA TCA TGG GAA AG |
237 |
Fang et al. (2014) |
|
bla TEM
|
CGC CGC ATA CAC TAT TCT CAG AAT GA ACG CTC ACC GGC TCC AGA TTT AT |
455 |
Monstein et al. (2007) |
|
bla CTX-M |
ATG TGC AGY ACC AGT AAR GTK ATG GC TGG GTR AAR TAR GTS ACC AGA AYC AGC GG |
593 |
Boyd et al. (2004) |
|
bla OXA
|
ACA CAA TAC ATA TCA ACT TCG C AGT GTG TTT AGA ATG GTG ATC |
813 |
Ouelletteet al. (1987) |
| PABLs encoding gene | |||
| CMY II |
AGCGATCCGGTCACGAAATA CCCGTTTTATG CACCCATGA |
695 |
Junyoung et al. (2009) |
| Colistin resistance encoding gene | |||
| mcr1 |
AGTCCGTTTGTTCTTGTGGC AGATCCTTGGTCTCGGCTTG |
320 |
Ana Rita Rebelo et al. (2018) |
Esc Resistant E. Coli Isolates Identification
ESC E. coli isolates were determined by resistance to one or more third and fourth generation cephalosporins (CDC, 2020).
Antimicrobial Susceptibility Test
Antibiotic susceptibility test of E. coli isolates against nine different antibiotics was performed according to the Kirby-Bauer disc diffusion method using Mueller-Hinton agar (Bauer et al., 1966). The susceptibility of the E. coli isolates against each antimicrobial agent was measured and readings have been noted and compared with the Clinical and Laboratory Standards Institute guidelines (CLSI, 2020) (Table 2).
Table 2: Different antimicrobials used in disc diffusion method.
| Antibiotic | Concentration | Abbreviation |
| Tetracycline |
30 μg |
TE |
| Cefoxitin |
30 μg |
CX/FOX |
| Cefotaxime |
30 μg |
CTX |
| Trimethoprim Sulfamethoxazole |
1.25/23.75 μg |
SXT |
| amoxycillin clavulanate |
20/10 μg |
AMC |
| Cefquinome |
30 μg |
CEQ |
| Cetazidime |
30 μg |
CAZ |
RESULTS AND DISCUSSION
A high phenotypic prevalence (using EMB) of E. coli intramammary infection from mastitic dairy cows (68 out of 205) at the percent of 33%, where the genotypic prevalence (PCR to detect 16S rRNA gene) revealed 30.7% (63 out of 205) of dairy cows contract E. coli infection. The prevalence rate of bovine mastitis caused by E. coli was 33% of the overall milk samples. Most infections of the cows with E. coli are from their environment, as faces and straw as hypothesized by Lipman et al. (1995).
The proved 63 E. coli strains were then subjected for detection of E. coli O157 virulence gene, where only two STEC strains having rfbEO157 encoding gene had been detected using uniplex PCR at a percentage of 3.2% (Figure 1 & 2). Shiga toxin-producing E. coli (STEC) strains considered to be the most important pathogens of a recently emerged group of food-borne strains in the milk of infected cows. This type of strain has been associated with outbreaks of diarrhoea, gastroenteritis and hemorrhagic colitis (HC) or the hemolytic uremic syndrome (HUS) in humans (Karmali, 1989; Paton and Paton, 1998; Beutin et al., 2004). It is agreed with Hassan et al. (2012) who recorded that STEC strains can induce bovine mastitis and reduce milk quality for human consumption because some of the mastitis cases are subclinical and the diagnosis is based solely on accurate diagnostic tests.
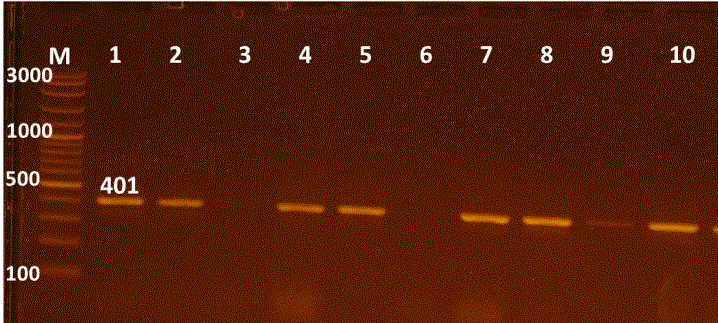
Figure 1: Uniplex PCR for 16s rRNA detection of E. coli, Lane M:100-3000bpDNA marker; Lanes 1-10 were positive E. coli isolates.
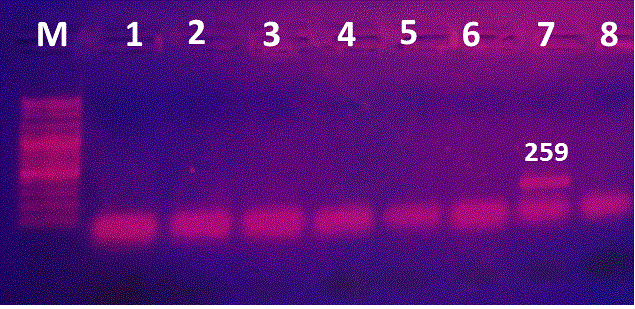
Figure 2: PCR for detection of rfbEO157 encoding gene. Lane M:100-3000bpDNA marker; Lane 1, 2: positive isolate for rfbEO157 gene; Lanes 3-7: negative strains.
Domestic ruminants, especially cattle, sheep and goats, are the principal reservoirs of STEC strains that cause human infections (Zschock et al., 2000; Chapman et al., 2001).
Regarding resistance genes, all of the isolates (100%) encoded TEM-type ESBLs, while none of which (0%) encoded OXA-type ESBLs. But, both CTX-M-type ESBLs and SHV-type β -lactamases were encoded in 53.9% (34 out of 63) and 4.7% (3 out of 63) of the ESBL isolates, respectively. Also, 27% exhibited CMYІІ-type PABLs. For plasmid-mediated colistin resistance encoding gene (mcr-1) was expressed in only one E. coli isolate at a percentage of 1.6% (1 out of 63). Regarding phenotypic non-β-lactams antimicrobial resistance, about 45.7% of E. coli isolates showed resistance to tetracycline, while 37.3% exhibited resistance to Trimethoprim-Sulfamethoxazole. This finding is similar to Sobhy et al. (2020) who associated higher resistance to Tetracyclines and Sulfamethoxazole/Trimethoprim with the prolonged use of these cheap antibiotics in the Egyptian dairy farms. In the same regard, Okubo et al. (2019) reported about 47.8% of bovine E. coli strains were co‐resistant to Ampicillin, Tetracycline and Sulfamethoxazole/Trimethoprim due to extensive use of these antimicrobials in Ugandan livestock.
Concerning molecular detection of the mcr-1 gene in ESC E. coli isolates were about 3%. This finding is in contrast to Umpiérrez et al. (2017) who recorded the absence of the mcr-1 gene in bovine E. coli strains, while Haenni et al. (2016) who detected an increase in the proportion of mcr-1 within ESBL-producing E. coli strains ranged from 4.76% in 2006 to 21.28% in 2014, prompting reducing colistin exposure.
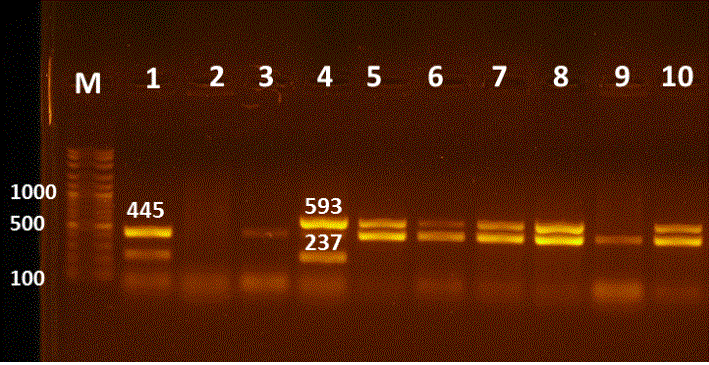
Figure 3: Multiplex PCR for detection of blaTEM, blaCTXM, blaSHV and blaOXA genes in E. coli isolates. Lane M:100-3000bpDNA marker; Lane 1 positive blaTEM& blaSHV at 445, 237bp, respectively; Lanes 3,9 positive blaTEM at 445bp; Lane 4 positive blaCTXM, blaSHV at 593, 237bp, respectively; Lanes 5-8&10 positive strains for blaTEM & blaCTXM at 445 and 593bp; Lane 2 negative sample.
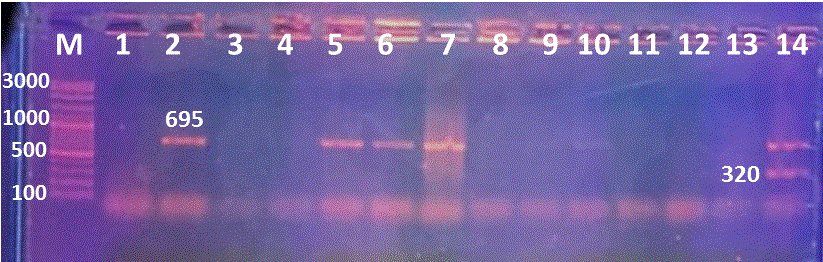
Figure 4: Multiplex PCR for mcr1 and blacmyІІ genes detection, Lane M:100-3000bpDNA marker; Lanes 2,5, 6 and 7 were positive blacmyІІ at 695bp, while Lane 14 positive mcr1 and blacmyІІ at 320, 695bp, respectively. Lanes 1,3, 4 and 8-13 are negative samples.
21 E. coli isolates demonstrated phenotypic resistance to cefotaxime (CTX) and were encoding for the blaCTX gene. But, only 4 isolates showed phenotypic resistance although they lack blaCTX resistance gene (Table 3 & 4) & (Figure 3 & 4). The phenotypic resistance to ESCs antibiotics as (Cefotaxime, Cefquinome and Ceftazidime) was increased, due to their extensive and widespread use in veterinary medicine as mentioned by Ahmed and Shimamoto, (2015) when they declared that ESCs (3rd and 4th generation Cephalosporins) are necessary antibiotics used in vet
Table 3: Comparison between phenotypic and genotypic antimicrobial resistance pattern of E. coli isolates and their relation to O157 virulence gene.
| Isolate code | O157 | Resistance phenotype | ESBLs genes |
Colistin resistance mcr1 |
PABLs blacmyІІ |
|||||||||
|
Β-lactams |
Non Β-lactams |
|||||||||||||
| CX | CAZ | CTX | CEQ | AMC | SXT | TE |
blaTEM |
blaCTX |
blaSHV |
blaOXA |
||||
| 2, 18 | + | + | + | + | + | |||||||||
| 11, 57 | + | + | ||||||||||||
| 14 | + | + | + | + | + | |||||||||
| 15 | + | + | + | + | + | |||||||||
| 16 | + | + | + | |||||||||||
| 17 | + |
+ |
+ | + | + | |||||||||
| 20, 175, 177, 178 | + | + | + | |||||||||||
| 22, 50, 35 | + | + | + | + | + | + | ||||||||
| 23 | + | + | ||||||||||||
| 28, 41, 187, 204 | + | + | ||||||||||||
| 34, 46 | + | + | + | |||||||||||
| 42 | + | + | + | + | + | + | ||||||||
| 45 | + | + | + | + | + | |||||||||
| 48, 188 | + |
+ |
+ | + | ||||||||||
| 81 | + | + | + | + | + | + | + | + | ||||||
| 103, 104, 109, 111 | + | + | + | |||||||||||
| 110, 202 | + | + | + | + | ||||||||||
| 130 | + | + | + | + | + | + | + | + | ||||||
| 148 | + | + | ||||||||||||
| 149, 150, 163 | + |
+ |
+ | |||||||||||
| 151 | + | + | + | |||||||||||
| 155 | + | + | ||||||||||||
| 157 | + | + | + | |||||||||||
| 176 | + | + | + | + | + |
+ |
+ | |||||||
| 179 | + | + | + | + | ||||||||||
| 181 | + |
+ |
+ | + | + | + | ||||||||
| 182 | + | + | + | + | + | |||||||||
| 183 | + | + | + | |||||||||||
| 184 | + |
+ |
+ | |||||||||||
| 185 | + | + | + | + | + | |||||||||
| 186 | + | + | + | + | + | + | + | + | ||||||
| 189 | + | + | + | + | + | + | + |
+ |
+ | + | ||||
| 191 | + | + | + | + | + | + | + | |||||||
| 192 | + | + | + | + | + | + | + | + | + | + | ||||
| 194 | + | + | + | + | + | |||||||||
| 195 | + | + | + | + | + | + | + | + | + | |||||
| 196 | + | + | + | + | + | + | + | + | + | |||||
| 198, 200 | + | + | ||||||||||||
| 203 | + | |||||||||||||
Table 4: Multidrug resistance pattern and resistance gene of E. coli isolates
| Number of isolates | Resistance phenotype |
16s RNA |
O157 | ESBLs genes |
Colistin (mcr1) |
PABLs (blacmyІІ) |
|||
| TEM | CTX | SHV | OXA | ||||||
| 17 | SXT, CTX, CEQ | + | - | + | + | - | - | - | - |
| 22, 50 | SXT, TE, CTX, CEQ | + | - | + | + | - | - | - | - |
| 35 | SXT, TE, CTX, CEQ | + | - | + | + | - | - | - |
- |
| 81 | SXT, TE, CAZ, CTX, CEQ | + | - | + | + | + | - | - | - |
| 14 | CX, CAZ | + | + | + | + | - | - | - |
- |
| 109, 111,104, 103, 77 | SXT, TE | + | - | + | - | - | - | - | - |
| 163, 150, 149 | CX, CAZ | + | - | + | - | - | - | - | - |
| 157 | TE, AMC | + | - | + | - | - | - | - | - |
| 151 | SXT, CX | + | - | + | - | - | - | - | - |
| 130 | SXT, TE, AMC, CAZ, CTX, CEQ | + | - | + | + | - | - | - | - |
| 110 | SXT, TE | + | - | + | - | - | - | - | + |
| 2, 18 | TE, CTX, CEQ | + | - | + | + | - | - | - | - |
| 42 | TE, CAZ, CTX, CEQ | + | - | + | + | - | - | - |
- |
| 45 | TE, CAZ, CTX | + | - | + | - | + | - | - | - |
| 15 | CAZ, CTX, CEQ | + | - | + | + | - | - | - | - |
| 48, 188 | CTX, CEQ | + | - | + | + | - | - | - | - |
| 181 | TE, CTX, CEQ | + | - | + | + | - | - | - | + |
| 182 | TE, CEQ | + | - | + | + | - | - | - | + |
| 185 | CTX, CEQ | + | - | + | + | - | - | - | + |
| 186 | SXT, TE, CAZ, CTX, CEQ | + | - | + | + | - | - | - | + |
| 189 | TE, CX, CAZ, CTX, CEQ | + | + | + | + | - | - | + | + |
| 195 | SXT, TE | + | - | + | - | - | - | - |
- |
Table 5: Extended spectrum and plasmid mediated ampicilin β-lactamases and colistin resistance genes of E. coli isolates from mastitic milk samples.
|
E. coli isolates |
ESBLs | PABLs no. | Colistin no. | ||||
|
blaTEM |
blaCTXM |
blaSHV |
blaOXA |
blaCMYІІ |
Mcr-1 | ||
| No. | 63 | 63 | 34 | 3 | 0 | 17 | 1 |
| % | - | 100 | 54 | 4.7 | 0 | 27 |
1.6 |
erinary and human medicine.
The molecular detection of resistance genes such as ESBLs, PABLs and colistin resistance genes revealed that all E. coli isolates harbour blaTEM and about half of them bear blaCTXM, while 27% of the isolated have blaCMYІІ and only one isolate (1.6%) has Mcr-1 (Table 5) (Figure 3 & 4). Chirila et al. (2017) and Poirel et al. (2018) declared that E. coli may develop resistance to antimicrobials by chromosomal genes mutation or by horizontal gene transfer of resistance genes within commensal and pathogenic E. coli strains, rendering E. coli as a major reservoir of resistant genes that could be responsible for human and veterinary treatment failure.
There was a significant increase in isolates with resistance genes and exhibit ESC resistance as isolates carried blaTEM, blaTEM+ blaCTXM and blaTEM+ blaCTXM+ blaCMYІІ. In addition, ESC susceptible isolates also bear resistance genes such as blaTEM+, blaTEM+ blaCTXM+ blaCMYІІ, blaTEM+ blaCTXM and blaTEM+ blaCMYІІ (Table 6). ESC resistant E. coli strains were determined according to their resistance to one or more of
Table 6: ESC resistant and susceptible E. coli isolates and their antimicrobial resistance genes profile.
| ESC resistance | No. of isolates |
blaTEM+ |
blaTEM+ |
blaTEM+& blaCTXM+ |
blaTEM+, blaCTXM+& blaCMYІІ+ |
|||||
|
blaCTXM |
blaSHV |
blaOXA |
blaCMYІІ |
Mcr-1 |
blaSHV |
blaCMYІІ |
Mcr-1 | |||
| Resistant | 33 |
15 (45.4%) |
1 (3%) |
- |
1 (3%) |
- |
7 (21.2%) |
1 (3%) |
7 (21.2%) |
1 (3%) |
| Susceptible | 26 |
5 (19.2%) |
1 (3.8%) |
- |
3 (11.5%) |
- |
12 (46%) |
- |
5 (19.2%) |
- |
Table 7: Antibiotic susceptibility profile of E. coli isolates
| Antimicrobial agents | Abbreviation |
Conc. (µg) |
Susceptible | Intermediate | Resistance | ||||
| No. | % | No. | % | No. | % | ||||
|
Non β-lactams |
Trimethoprim/Sulfamethoxazole | SXT | 1.25/23.75 | 38/59 | 64.4 | 0/59 | 0 | 22/59 | 37.3 |
| Tetracycline | TE | 30 | 26/59 | 44.1 | 7/59 | 11.8 | 27/59 | 45.7 | |
|
β-lactams |
Cefoxitin | CX | 30 | 48/60 | 80 | 3/60 | 5 | 9/60 | 15 |
| Amoxicillin/ clavulanate | AMC | 20/10 | 50/59 | 84.7 | 4/59 | 6.7 | 6/59 | 10.1 | |
| Ceftazidime | CAZ | 30 | 35/59 | 59.3 | 6/59 | 10.2 | 19/59 | 32.2 | |
| Cefotaxime | CTX | 30 | 32/59 | 54.2 | 3/59 | 5.1 | 25/59 | 42.3 | |
| Cefquinome | CEQ | 30 | 29/59 | 49.1 | 6/59 | 10.2 | 25/59 |
42.3 |
|
the 3rd and 4th generations of cephalosporins (CDC, 2020). In this line, the percentage of ESC resistant isolates was high 56% (33 out of 59). Based on ESCs resistance pattern and the presence of antimicrobial resistance genes, it was observed that the highest percentage of ESCs resistant E. coli isolates had blaTEM+ blaCTXM followed by blaTEM+ and blaTEM+ blaCTXM+ blaCMYІІ. Although ESC susceptible E. coli isolates possess β-lactamase resistance genes but were not expressed in vitro or phenotypically. It is agreed with Ahmed et al. (2009) when declared that E. coli strains showed phenotypic antibiotic multi-resistance primarily against ESCs, including Cefotaxime and Ceftriaxone, and other non-β-lactams; especially Tetracycline, Sulfamethoxazole/ Trimethoprim, Nalidixic acid and Ciprofloxacin.
With regard to E. coli isolates, about 45.4% of ESC resistant isolates were attributed to the presence of blaTEM and blaCTXM genes followed by blaTEM and blaTEM+ blaCTXM+ blaCMYІІ combinations at a rate of 21.2% for each. Almost all ESBLs producing isolates having blaTEM and two combinations including blaTEM + blaCTXM + blaCMYІІ and blaTEM + blaCTXM + blaCMYІІ + mcr1, which conferred resistance to Cefquinome as ESCs. It was contrary to Filioussis et al. (2020) who determined ESBLs producing strains from milk have blaTEM + blaSHV + mcr-1 and blaTEM + blaCTXM + mcr-1 combinations that conferred resistance to ESCs especially cefquinome.
Most ESC resistant E. coli isolates 26/33 (78.8%) have more than one antimicrobial resistance gene. This was agreed with Awosile et al. (2018) who determined the existence of two or more β-lactamase genes within 44% of ESC resistant strains, illustrating the phenotypic resistance of E. coli isolates is highly dependent on the co-existence of two or more β-lactamase genes in such isolates. The majority of ESC E. coli isolates 25/33 (75.7%) have β-lactamase CTX as reported by Livermore et al. (2007) and Seiffert et al. (2013) when it was noted that the worldwide evolution of β-lactamase CTX (Cefotaximase) has been identified and is known to be the most common cause of ESC resistance in the Enterobacteriaceae.
Concerning the antimicrobial susceptibility profile; Seven E. coli isolates (11.8%) were susceptible to all antimicrobials and only two isolates (3.4%) were resistant to all antimicrobials, eleven isolates (18.6%) expressed resistance to a single compound, and 36 isolates (61%) showed resistance to more than one antimicrobial agent. Whereas, twelve isolates (20.3%) were expressing resistance to 3 related compounds (i.e., Cefotaxime, Cefquinome and Ceftazidime) as extended spectrum cephalosporins (Table 7) & (Figure 5). Multidrug resistance (MDR) among E. coli isolates was high, where 61% of E. coli isolates (36/59) showed MDR against two or more antimicrobials, nine isolates (15.3%) exhibit MDR for β-lactam antibiotics, six isolates (10.2%) for non-β-lactam antibiotics, 19 isolates (32.2%) were resistant to both (β-lactams and non-β-lactams) and two isolates (3.4%) showed resistance to all antimicrobial used in this study. Our results were compatible with Pasayo et al. (2019) who declared that frequent use of antibiotic treatment leading to the production of multi-resistant strains that pose a major public health threat. But not aligned with Ahmed et al. (2009) who stated that lower multi-antibiotic resistance has been found in E. coli strains are 10.4%.
CONCLUSION
The emergence of antimicrobial resistance in particular to the recently introduced antimicrobials such as 3rd and 4th generations of cephalosporins in E. coli strains attributed to antimicrobial misuse in dairy farms for bovine mastitis therapy. E. coli strains acquire antimicrobial resistance through plasmid-mediated transfer leading to a widespread of multidrug resistance to ESCs, β-lactams and non-β-lactams antibiotics that can induce treatment failure in dairy farms.
ACKNOWLEDGMENTS
The authors would like to thank Dr. Mohamed Ibrahim, farm managers, staff and all helped with this work.
CONFLICT OF INTEREST
There are no conflicts of interest in this report, according to both of the contributors.
AUTHOR CONTRIBUTIONS
Adel Abdel-Azim Fayed approved of and arranged the tests. Soliman Mohamed Soliman created the hypothesis, computed it, and analyzed the results. The experiment was carried out by Samah El-sayed Mahmoud, and the manuscript was written with input and help from all contributors. Adel Abdel-Azim Fayed and Samia Abd El-Hamid Ahmed created the model and nearly all of the technical data. All writers discussed the conclusions, offered critical input, assisted in the creation of the study, and collaborated on the final manuscript.
ETHICS STATEMENT
All research procedures were carried out in accordance with the Animal Ethics Committee of APRI, ARC, Egypt’s recommendations for the treatment and use of lab animals.
REFERENCES