Advances in Animal and Veterinary Sciences
Research Article
Phenotypic, Genotypic Resistance and Virulotyping of Staphylococcus aureus Isolated from Ready-to-Eat Food in Egypt
Alaa Eldin M.A. Morshdy1, Mohamed A. Hussein1, Abdallah M.A. Merwad2*, Hanan, M. El. Lawendy3, Afaf H. Mohamed3
1Department of Food Control, Faculty of Veterinary Medicine, Zagazig University, Egypt; 2Department of Zoonoses, Faculty of Veterinary Medicine, Zagazig University, Egypt; 3Department of Food Hygiene, Animal Health Research Institute, Zagazig, Provential Laboratory, Egypt.
Abstract | The present study aimed to determine phenotypic and genotypic characterization of antimicrobial resistance and some virulent genes in Staphylococcus aureus (S. aureus) isolated from ready- to- eat (RTE) food purchased from commercial restaurants at Sharkia, Egypt. One hundred RTE food including kofta, shawarma, sausage and fried shrimp were streaked on Baired-Parker agar plates followed by biochemical identification of isolates. Twenty seven S. aureus isolates were screened for antibiotic resistance by disk diffusion test. Uniplex PCR was used to detect the antimicrobial resistance genes for methicillin (mecA), beta-lactamase (blaZ), aminoglycoside [aac (6’) aph(2’’)] and macrolides (mph C) and virulence genes (spa and clfA) in four phenotypically multidrug resistant (MDR) S. aureus isolates. Furthermore, enterotoxin genes were screened in the four isolates of S. aureus using multiplex PCR. In this study, S. aureus strains showed highest resistance to methicillin and cefotaxime (100%) followed by amoxicillin (85.18%) and gentamicin (81.48%). The multiple antibiotic resistance (MAR) index of S. aureushad a range of 0.36 to 0.9 with an average 0.58. Three (25%) isolates from kofta were multi-resistant to 10 antibiotics and one strain (9.09%) of S. aureus from shawarma was resistant to 10 antimicrobials. The four isolates of phenotypically resistant S. aureus to methicillin, amoxicillin and gentamicin were positive (100%) for each of mecA, blaZ and aac (6’) aph (2’’) resistance genes; while, two isolates showed resistance to macrolides were positive for mphC gene. The four multidrug resistant (MDR) S. aureus were negative for any enterotoxin genes. Additionally, all examined MDR isolates of S. aureus from RTE food harboring spa and clfA genes (100%). The higher contamination of different RTE food with MDR S. aureus indicates improper hygienic measures. Also, the higher MAR index of S. aureus isolates has high risk potential for consumers.
Keywords | Phenotypic resistance, Staphylococcus aureus, Ready-to-Eat Food, Virulence genes, Antimicrobial resistance genes
Received | September 19, 2019; Accepted | October 26, 2019; Published | December 12, 2019
*Correspondence | Abdallah M.A. Merwad, Department of Zoonoses, Faculty of Veterinary Medicine, Zagazig University, Egypt; Email: merwad.abdallah@yahoo.com
Citation | Morshdy AEMA, Hussein MA, Merwad AMA, Hanan, El Lawendy M, Mohamed AH (2019). Phenotypic, genotypic resistance and virulotyping of staphylococcus aureus isolated from ready-to-eat food in Egypt. Adv. Anim. Vet. Sci. 7(s2): 63-70.
DOI | http://dx.doi.org/10.17582/journal.aavs/2019/7.s2.63.70
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2019 Morshdy et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
Staphylococcus aureus (S. aureus) is considered as one of the major food borne bacterial pathogens recovered from RTE meat and causing several food poising outbreaks (Diep et al., 2006). This pathogen have the ability to grow within temperature range of 15-45 ºC but requires higher concentration (10%) of NaCl (Behling et al., 2010). S. aureus could multiply quickly at the room temperature and release toxins that induce illness. It has been estimated that S. aureus responsible for 241,000 cases of foodborne illness each year in United States of America (Wu et al., 2018). This bacterium could be detected in meat (Jackson et al., 2013) and fish (Saito et al., 2011). The contamination of food with S. aureus could be resulted from inadequate handling, improper conditions of storage, inadequate hygienic measures and post-production microbial contaminations (Ray, 2001). The presence of S. aureus as one of microbial contaminants in RTE meathas created public health hazard and food safety concerns (Mehndiratta and Bhalla, 2014), due to their pathogenicity and zoonotic importance (Razmyar et al., 2017). The antimicrobial susceptibility testing of S. aureus isolates recovered from RTE meat and retail chicken were studied in Egypt (Morshdy et al., 2018b) and India (Laxminarayan and Chaudhury, 2016).
Methicillin-resistant S. aureus (MRSA)occurs in humans and food of animal origin due to acquirement of genes that encode penicillin-bindingprotein-PBP2’ or PBP2a (Chen et al., 2009). Besides, a divergent homologue of the mecA gene (mec C) verified 70% homology to the methicillin resistance (mec A) gene (Garcia-Alvarez et al., 2011). Also, frequent contamination of retail meat and poultry occur with MDRS. aureus; and 96%of S. aureus strains showed resistance to at least oneantibiotic and 32% of susceptibility profiles were found in S. aureus isolates with several resistance to multiple antibiotic classes (Waters et al., 2011). Some studies highlighted the role of meat and poultry products in inducing human infection and colonization (de Jonge et al., 2010). The majority of virulence factors of S. aureus are cell surface protein including matrix adhesion molecules which mediates bacterial binding to the host tissue and extracellular matrix including collagen and fibrinogen (Foster et al., 2014). The components of S. aureus surface include an elastin binding protein and clumping factors A andB (clfA, clfB) (Atshan et al., 2012). S. aureus surface proteins are critical for success of the bacteria as a commensal organism and also as a pathogen (Foster et al., 2014). Staphylococcal protein A (SPA), another important virulence factor, is a multifunctional surface protein located on FC fragment of IgG of mammalian species inducing immune evasion and could act as super antigen of B-cell (Foster et al., 2014; Sutra and Poutrel, 1994). Staphylococcal food poisoning is associated with presence of staphylococcal enterotoxins (SEs) (Le Loir et al., 2003). These enterotoxins are produced in the exponential phase of S. aureus growth. They causes food poisoning in human when the count of S. aureus reaches at least 105 -108 CFU/g in food (Seo and Bohach, 2003). Therefore, the aim of current study was to determine the phenotypic resistance and genotypic characterization of antimicrobial resistance genes, some virulence genes (clf A and spa) and enterotoxin genesin S. aureus strains recovered from some RTE food (kofta, shawarma, sausage and fried shrimp) purchased from restaurants in SharkiaGovernorate, Egypt.
MATERIALS AND METHODS
COLLECTION OF FOOD SAMPLES
One hundred RTE food samples comprising of kofta, shawrma, sausage and fried shrimp (25, each) were purchased from commercial restaurants and supermarkets at Sharkia Governorate, Egypt during the period from October, 2017 to April, 2018. The collected samples were packed into sterile lunch boxes. The samples of RTE food were directly placed in a cooler box; and then directly transferred to the Department of Food Control, Faculty of Veterinary Medicine, Zagazig University, and stored at 4 °C till analysis. All samples were microbiologically analyzed within 4 hrs of purchase.
PROCESSING OF RTE FOOD SAMPLES
The food samples were prepared according to the previously described method (APHA, 1992). Twenty five gram of each RTE food samples were added to a volume of 225 ml sterile buffered peptone water (Oxoid, CM509, UK). The mixture was homogenized by a stomacher for 1 min, and then the homogenate was transferred in a sterile test tube. Afterwards, the homogenate (1 ml) was placed in sterile test tube containing0.1% peptone water (9 ml), and then subjected to 10-fold serial dilution up to 107.
ISOLATION AND IDENTIFICATION
The isolation procedure of S. aureus was carried out according to the previously described method (Thapaliya et al., 2017). The prepared serial dilution (0.1 ml) was streaked on Baired–Parker agar plate (Oxoid, CM0275, UK) and incubated at 37 ºC for 24 hrs. The suspected colonies are black-colord surrounded by a wide clear area with opacity. The presumptive coloniesshowing positive egg yolk reaction were subjected to biochemical identification. The motility test (ICMSF, 1996), catalase activity test (Macfaddin, 2000) and coagulase activity test (APHA, 1992) were carried out as described before. The colonies revealing coagulase positive, Gram positive cocci, catalase positive were biochemically distinguished as S. aureus.
ANTIMICROBIAL SUSCEPTIBILITY TESTING
The biochemically identified isolates of S. aureus were tested for their antimicrobial susceptibility against eleven antibiotics on Mueller-Hinton agar plates (Oxoid, England) using the Kirby Bauer disk diffusion test (Bauer et al., 1966). The antimicrobial panels and their disc concentrations including methicillin (ME; 10 μg), cefotaxime (CTX, 30μg); Amoxicillin-clavulanic acid (AMC, 30μg), gentamicin (CN, 10µg), erythromycin (E, 15μg), clindamycin (DA, 2μg), doxycycline (DO, 5μg), ciprofloxacin (CIP, 5μg), sulfamethoxazole-trimethoprim (SXT, 25µg), chloramphenicol (C, 30µg) and vancomycin (VA, 30μg) were obtained from Thermo scientific, Oxoid, USA. Interpretation of the results was carried out according to the standards of Clinical Laboratory Standard Institute guidelines (CLSI, 2016). The multiple antibiotic resistance index of S.aureus isolate was calculated according to the formula stipulated by (Singh et al., 2010) as the following equation:
MAR index = Number of antibiotics with resistance profile / the number of used antibiotics
Molecular Detection Of Antimicrobial Resistance And Virulence Genes
The genomic DNA was extracted from four biochemically identified S. aureus isolates with phenotypic resistance and isolated from sharwma, sausage, fried shrimp and kofta using QIAamp DNA mini kit (Takara Kit, Catalogue no.51304, Japan). The selected primer sequences and cycling conditions were carried out to amplify the resistance genes to methicillin (mecA) (McClure et al., 2006), beta-lactamase (bla Z) and aminoglycoside [aac(6’)aph(2’’)](Duran et al., 2012) and macrolide (mph C) (Schlegelova et al., 2008) in S. aureus isolates using uniplex PCR (Table 1).
The two virulence factors including staphylococcal protein A (spa gene) (Wada et al., 2010) and clumping factor A (clf A gene) (Mason et al., 2001) were molecularly detected in S. aureus isolates using uniplex PCR. Also, the oligonucletides sequences and PCR cycling conditions to identify enterotoxin genes (sea, seb, sec, sed and see) in S. aureus were performed by multiplex PCR according to (Mehrotra et al., 2000) (Table 1). The electrophoresis of PCR products was performed on 1.5% agarose gel having ethidium bromide (0.5 μg/ml) with gel pilot 100 bp ladder (QIAGEN, USA). Positive control isolate of S. aureus was included in each reaction, while Escherichia coli isolate was used as negative one.
RESULTS AND DISCUSSION
S. aureus is one of the most important food borne pathogens in RTE food products and is incriminated in different infections all over the world (Diep et al., 2006). Additionally, this bacterium is responsible for 12.5% of food borne bacterial outbreaks in China and is considered the third most frequently pathogen after Vibrio parahaemolyticus outbreaks (27.8%) and Salmonella food poisoning (23.1%) (Wei-Wei et al., 2018). S. aureus could adapt to different environmental conditions and quickly acquire resistance to virtually all antimicrobials (McCallum et al., 2010). Recently, MDR S. aureus were recorded in outbreaks of food poisoning that were related with consumption of food products (Papadopoulos et al., 2018). Our study showed that the incidence of S. aureus in RTE food was 27%. However, S. aureuswas recoverd with lower percentage (17.9%) from RTE food in Taiwan (Fang et al., 2003). In the present study, the highest isolation percentage of S. aureus (48%, 12/25) was detected in kofta sandwiches, followed by shwarma sandwiches (36%, 9/25); while the lowest incidence (12%, 3/25) was recovered from sandwiches of sausage and fried shrimp. On the contrary,lower isolation percentages of S. aureus was recovered in minced meat (10%), sausage (8%) and beef burger (10%) in Egypt (Morshdy et al., 2018a). In Egypt, higher incidence of S. aureus was detected in examined kofta (90%), luncheon (50%), burger (75%), shawarma (80%), hawawshi (65%), and sausage sandwiches (60%) (Morshdy et al., 2018b). These variations could be attributed to difference in the system of cooking (frying, roasting and grilling), the microbiological quality of raw ingredients, size and shape of meat pieces, cooking utensils (oven, grill, stew pot) and type of seasoning ingredients (sauces, spices, vegetables) (Daelman et al., 2013).
In the current study, the phenotypic resistance of 27 isolates of S. aureus from RTE food comprising kofta (12 isolates), shawrma (9), sausage (3) and fried shrimp (3) were investigated against 11 antimicrobials using the disc diffusion method (Table 2). In our study, isolates of S. aureus clarified the peak resistance to methicillin and cefotaxime (100%) followed by amoxicillin (85.18%) and gentamicin (81.48%). Similarly, the resistance percentages of S. aureus from RTE meat products to methicillin and amoxicillin were 100% and 57.1%, respectively in Egypt (Morshdy et al., 2018a). The moderate resistance of S. aureus isolates was detected forerythromycin and clindamycin (62.96%), doxycycline (59.25%) and ciprofloxacin (48.14%); while the lower resistance of S. aureus isolates was found tosulfamethoxazole-trimethoprim (29.62%) and chloramphenicol (25.92%). Another study in Egypt investigating MDRS. aureus isolates derived from RTE meat sandwitches, and the results demonstrated that 84% of S. aureus isolates were resistant to erythromycin and 64% for oxytetracycline and sulphamethoxazol (Morshdy et al., 2018b). However, erythromycin showed moderate resistance (52.7%) for S. aureus isolates recovered from retail meat and meat products in China followed by clindamycin (29.6%), gentammicin (19.4%), ciprofloxacin (16.9%) and amoxicillin/clavulanic acid (11%) (Wu et al., 2018). Our finding showed that 27S. aureusisolatesfrom RTE food were sensitive (100%) to vancomycin as listed in Table 2. However, resistance of S. aureus to vancomycin was found in 27.6% of camel meat and 54.5% of human hand swabs in Egypt (Al-Amery et al., 2019). In the current study, maximum resistance (100 %) of S. aureus isolates derived from RTE food were found to methicillin as MRSAcould be transmitted to cooked food products owing to temperature abuse during the storage and thus resulting in multiplication of MRSA strains. Moreover, the higher MRSA contamination of different RTE meat and chicken products may be possible associated with utilization of antibiotics in animal husbandry (de Boer et al., 2009). Also, the major reason for high resistant S. aureus strains could be related with supplements and growth promoters for animal feed and for human treatments. The difference in resistance percentage of S. aureus strains to antimicrobials could be accounted for size of sample, geographic area and period of sample collection (Ge et al., 2017).
Table 1: Primer sequences and PCR conditions for antimicrobial resistance genes, enterotoxin genes and virulence determinants in S. aureus strains derived from ready to eat food.
| Reference | PCR cycling conditions | Length of amplified product (bp) | Primer sequence (5'-3') | Gene |
|
(McClure et al., 2006) |
94˚C-5min;35cycles (94˚C-30s, 50˚C- 30s, 72˚C-30s);72˚C-7 min | 310 | GTA GAA ATG ACT GAA CGT CCG ATA A | mecA |
| CCA ATT CCA CAT TGT TTC GGT CTA A | ||||
|
(Duran et al., 2012) |
94˚C-5min; 35cycles (94˚C-30s, 54˚C- 30 s,72˚C-30s); 72˚C-7 min | 173 | ACTTCAACACCTGCTGCTTTC | blaZ |
| TGACCACTTTTATCAGCAACC | ||||
| 94˚C-5min;35cycles (94˚C-30s, 50˚C- 40 s, 72˚C-45s); 72˚C-10 min | 491 | GAAGTACGCAGAAGAGA | aac(6')aph(2'') | |
| ACATGGCAAGCTCTAGGA | ||||
|
(Mehrotra et al., 2000) |
94˚C-5min;35cycles (94˚C-30s,50˚C- 40 s,72˚C-45s);72˚C-10 min |
102 | GGTTATCAATGTGCGGGTGG | sea |
| CGGCACTTTTTTCTCTTCGG | ||||
| 164 | GTATGGTGGTGTAACTGAGC | seb | ||
| CCAAATAGTGACGAGTTAGG | ||||
| 451 | AGATGAAGTAGTTGATGTGTATGG | sec | ||
| CACACTTTTAGAATCAACCG | ||||
| 278 | CCAATAATAGGAGAAAATAAAAG | sed | ||
| ATTGGTATTTTTTTTCGTTC | ||||
| 209 | AGGTTTTTTCACAGGTCATCC | see | ||
| CTTTTTTTTCTTCGGTCAATC | ||||
|
(Schlegelova et al., 2008) |
94˚C-5min;35cycles(94˚C-30s,55˚C- 40 s,72˚C-45s); 72˚C-10 min | 722 | GAGACTACCAAGAAGACCTGACG | mphC |
| CATACGCCGATTCTCCTGAT | ||||
|
(Wada et al., 2010) |
94˚C-5min; 35cycles (94˚C-30s, 55 ˚C- 30 s, 72˚C-30s); 72 ˚C-7 min | 226 | TCA AGCA AAG AAC AAC AAA ATG C | spa |
| GCT TTC GGT GCT TGA GAT TC | ||||
|
(Mason et al., 2001) |
94˚C-5min; 35cycles (94˚C-30s, 55˚C- 40 s, 72˚C-45s); 72˚C-10 min | 638 | GCAAAATCCAGCACAACAGGAAACGA | clfA |
| CTTGATCTCCAGCCATAATTGGTGG |
Table 2: Antimicrobial susceptibility of 27 S. aureus isolates derived from ready to eat food by disc diffusion method.
| Antimicrobials (disc concentration/µg) | R | S | I |
| ME (10) | 27 (100) | 0 (0.00) | 0 (0.00) |
| CTX (30) | 27 (100) | 0 (0.00) | 0 (0.00) |
| AMC (30) | (85.18) | 4(14.81) | 0 (0.00) |
| CN (10) | 22 (81.48) | 2 (7.40) | 3(11.11) |
| E (15) | 17 (62.96) | 5 (18.51) | 5(18.51) |
| DA (2) | 17(62.96) | 9 (33.33) | 1(3.70) |
| DO (5) | 16(59.25) | 10(37.03) | 1(3.70) |
| CIP (5) | 13(48.14) | 10 (37.03) | 4(14.81) |
| SXT (25) | 8(29.62) | 9 (33.33) | 10 (37.03) |
| C (30) | 7(25.92) | 10(37.03) | 10(37.03) |
| VA (30) | 0 (0.00) | 27(100) | 0 (0.00) |
R: resistant; I: intermediate; S: sensitive; Data were represented by No (%); ME: methicillin; CTX: cefotaxime; AMC: Amoxicillin- clavulanic acid; CN: gentamicin; E: erythromycin; DA: clindamycin; DO: doxycycline; CIP: ciprofloxacin; SXT: sulfamethoxazole-trimethoprim; C: chloramphenicol; VA: vancomycin.
In this study, the range of MAR index for S. aureus was 0.36 to 0.9 with an average 0.58 as listed in Table 3. Moreover, 3 (25%) isolates of kofta origin were multi-resistant to 10 antibiotics, while one isolate (9.09) derived from shawrma was multi drug resistant to 10 antimicrobials and other two S. aureus strains (22.2%) originated from sharwma were resistant to 9 antibiotics (Table 3). Similarly, the average MAR index of S. aureus isolates was 0.59 from RTE sandwiches in Egypt (Morshdy et al., 2018b). In another study, 95.65% of S. aureus from pork meat in India have MAR index above 0.3 (Savariraj et al., 2019). The difference in the MAR index could be attributed to variations in the sources of food samples and geographic distributions, which had differential selective pressures for levels of antibiotic resistance and the methodologies of test (Robert-Pillot et al., 2014). In the current study, S. aureus strains with higher value of MAR index (>0.2) reflected high risk sources and poses high risk hazard for consumers; where they eat those RTE foods contaminated with such strains as previously supported (Robert-Pillot et al., 2014). The highest MAR index
Table 3: Antimicrobial resistance profiles and MAR index of S. aureus isolates derived from ready to eat food.
| MAR | Resistance profile | No. of isolates (%) | No. of resistant antibiotcs | Resistance Pattern | Ready to eat food |
| 0.9 | ME, CTX,AMC, CN,E,DA,DO,CIP,SXT,C | 3(25) | 10 | R1 |
Kofta (12 isolates) |
| 0.54 | ME,CTX,AMC,CN,E,DA | 6 (50) | 6 | R2 | |
| 0.45 | ME,CTX,CN,DA,DO | 2(16.7) | 5 | R3 | |
| 0.36 | ME, CTX,AMC, DO | 1(8.3) | 4 | R4 | |
| 0.9 | ME,CTX,AMC,CN,E,DA,DO,CIP,SXT, C | 1 (9.09) | 10 | R5 |
Shawrma
(9 isolates) |
| 0.81 | ME, CTX,AMC,E,DA,DO,CIP,SXT,C | 2(22.2) | 9 | R6 | |
| 0.72 | ME,CTX,AMC,DA,DO,CIP,SXT,C | 1(9.09) | 8 | R7 | |
| 0.63 | ME,CTX,CN,DA,CIP,SXT,DO | 1(9.09) | 7 | R8 | |
| 0.54 | ME,CTX,CN,E,DO,AMC | 1(9.09) | 6 | R9 | |
| 0.36 | ME, CTX,AMC,CN | 3(33.3) | 4 | R10 | |
| 0.54 | ME,CTX,AMC,CN,DO,CIP | 2(66.7) | 6 | R11 |
Sausage (3 isolates) |
| 0.45 | ME,CTX,AMC,CN,E | 1(33.3) | 5 | R12 | |
| 0.63 | ME,CTX,AMC,CN,E,DA,CIP | 1(33.3) | 7 | R13 |
Fried shrimp (3 isolates) |
| 0.54 | ME,CTX,AMC,E,DO,CIP | 2(66.7) | 6 | R14 | |
| 0.58 | Average | ||||
MAR: Multiple antibiotic resistance; ME: methicillin; CTX: cefotaxime; AMC: Amoxicillin-clavulanic acid, CN: gentamicin; E: erythromycin; DA: clindamycin; DO: doxycycline; CIP: ciprofloxacin; SXT: sulfamethoxazole-trimethoprim; C: chloramphenicol; VA: vancomycin.
in our study could be accounted for frequent exposure of human and farm animals to antibiotics in animals raised for food and for various purposes such as growth promotion and therapeutics and these resistant pathogens could be transmitted to humans through food (Nygard et al., 2008). This study indicated that MDR S. aureus should be included in the list of antimicrobial resistant pathogens which cause contamination of the food supply. Therefore, monitoring of antimicrobial resistance is necessary to determine the effectiveness of new generations of antibiotics and S. aureus isolates revealed resistance to antimicrobials owing to their capability to produce an exoplysaccharide barrier (Gündoğan et al., 2006) and carrying broad variety of MDR genes on plasmids which could be exchanged and spread to other species of Staphylococci (Neihart et al., 1988).
In the present study, the four strains of phenotypically resistant S. aureus to methicillin, amoxicillin and gentamicin were positive (100%) for mec A, bla Z and aac (6’) aph (2’’) genes with PCR products of 310, 173 and 491 bp, respectively (Figure 1). However, 2 out of 4 phenotypically resistant isolates to macrolide antibiotics (erythromycin) were positive for mphC gene with a PCR product of 722 bp (Figure 1). This finding agreed with previous reports of multidrug resistant S. aureus bearing mec A, bla Z and aac(6’)aph (2’’) from retailed meat products (Morshdy et al., 2018a), chicken meat and giblets (Darwish et al., 2018) in Egypt and RTE meat products (Xing et al., 2014) in China. Our study indicated that RTE meat, chicken and shrimp could be considered as important transmission route for antibiotic resistant S. aureus bearing multiple antibiotic resistance genes.
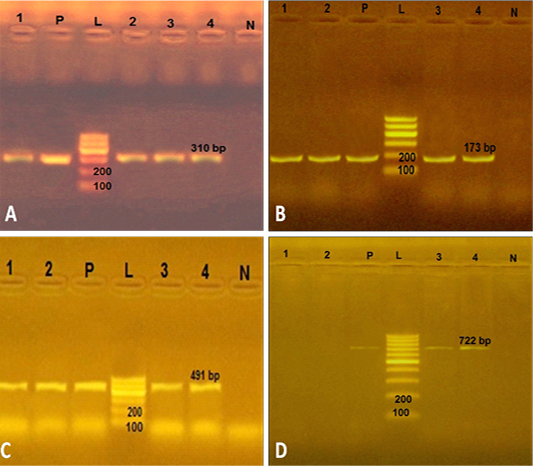
Figure 1: Uniplex PCR to amplify some antimicrobial resistance genes in 4 S. aureus strains from ready to eat food. A) PCR products of mec A gene. P: positive control isolate; N: negative control isolate of Escherichia coli; L: Ladder 100 bp; Lanes 1, 2, 3 and 4: positive strains for mec A gene (310 bp) isolated from kofta, shawrma, sausage and fried shrimp, respectively; B) PCR products of bla Z gene. The four S. aureus isolates (lanes1-4) were positive PCR product of 173 bp; C) PCR products of aac(6’)aph (2’’) gene. The lanes1-4 were with amplicon of 491 bp; D) PCR products of mph C gene. The two S. aureus isolates from kofta and shawarma (Lanes 1 and 2) were negative, while other two S. aureus strains from sausage and fried shrimp (lanes 3and4) were positive with PCR product of 722 bp.
In the current study, the 4 multidrug resistant S. aureus were negative for any enterotoxin genes. However, more than 74% of S. aureus strains derived from RTE food, processed meat and fish products in Bangladesh harbored enterotoxin genes with a distribution percentage of 26% for sea, 11% for seb, 49% for sec and 13% for sed genes (Islam et al., 2019). Furthermore all 4 multidrug resistant S. aureus from RTE food harboring the two virulence genes (spa and clf A genes) with PCR products of 226 and 638 bp, respectively (Figure 2). Conversely, lower distribution (76.92%) of clf A gene in S. aureus strains recovered from chicken meat in Iran (Momtaz et al., 2013). This study revealed that multidrug resistant S. aureus isolated from RTE food were MRSA isolates, possessed two virulence genes (clf A and spa genes), represent public health hazard for consumers.
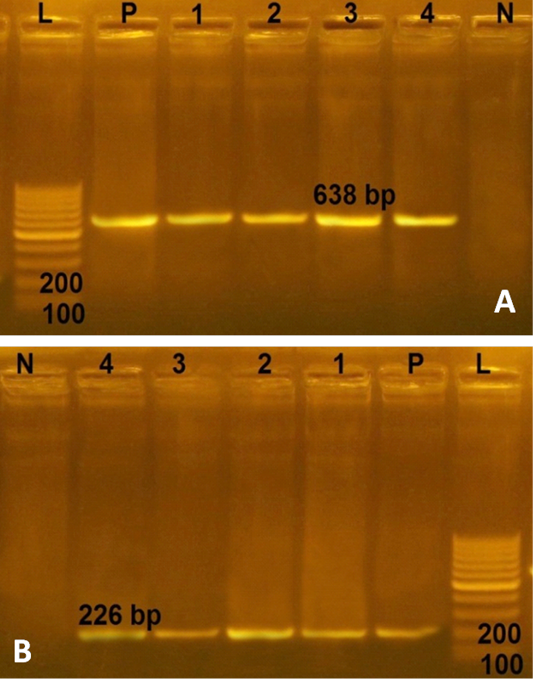
Figure 2: Agarose gel electrophoresies of two virulence genes (clf A and spa) in 4 strains of S. aureus isolated from ready to eat food. A) PCR products of clf A gene. P: positive control isolate; N: negative control; L: Ladder 100 bp; Lanes 1- 4: positive strains for clfA gene (638 bp) isolated from kofta, shawarma, sausage and fried shrimp, respectively; B) PCR products of spa gene. The four S. aureus isolates (lanes1-4) were positive with amplicons of 226 bp.
CONCLUSION
The higher contamination of different kofta, sharwma, sausage and fried shrimp as RTE food with multidrug resistant S. aureus indicates improper hygienic measures. Also, higher MRSA contamination of such RTE food could be associated with the wide use of antimicrobials in animal husbandry. Moreover, higher MAR index of S. aureus isolates poses high risk potential for consumers to eat such contaminated RTE food. Therefore, RTE food could be considered as important transmission route for phenotypic resistant S. aureus that carry resistance genes to methicillin, beta-lactamase, macrolide and aminoglycosides. Strict hygienic measures should be taken during the preparation of RTE food, besides strong legislations should be followed to produce food of high keeping quality.
AUTHORS CONTRIBUTION
All authors had contributed equally.
CONFLICT OF INTEREST
None of the authors have any conflict of interest to declare.
REFERENCES






