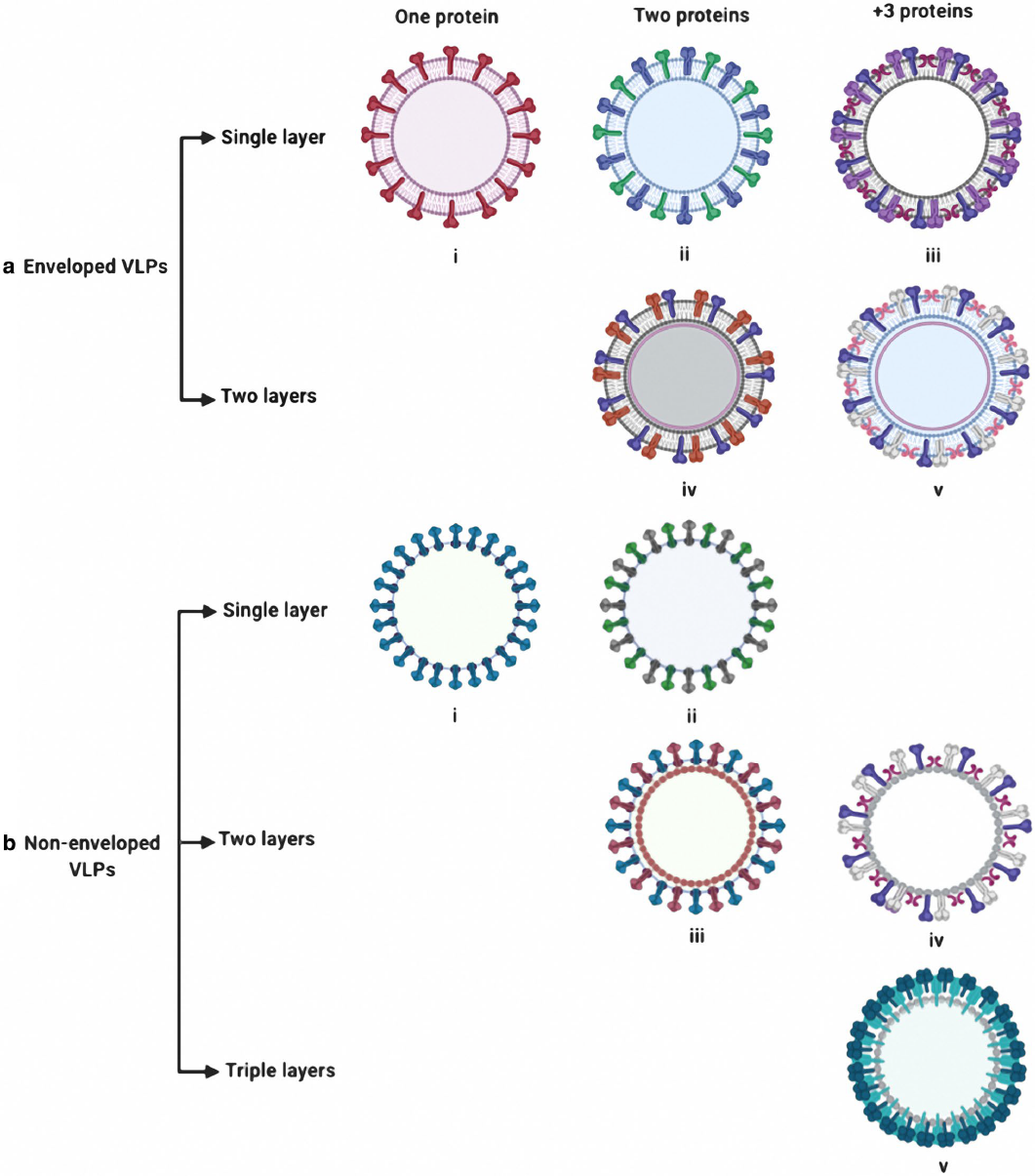
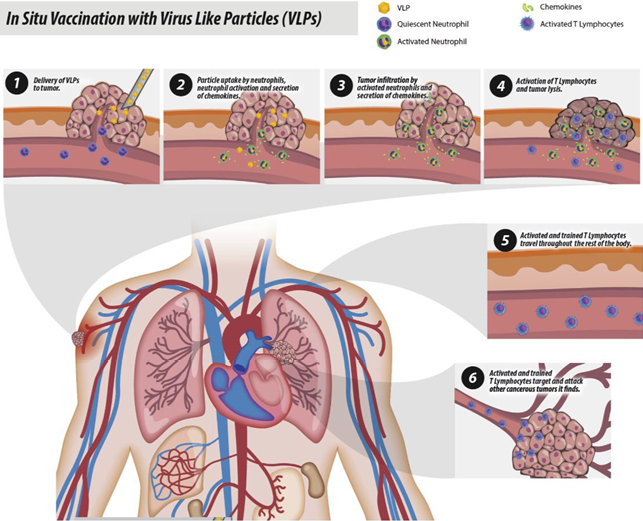
Virus Like Particles: Nanoparticles for Targeted Drug Delivery
Virus Like Particles: Nanoparticles for Targeted Drug Delivery
Khaled A. El-Dougdoug1, Wael S. El-Araby2* and Rehab, A. Dawoud3
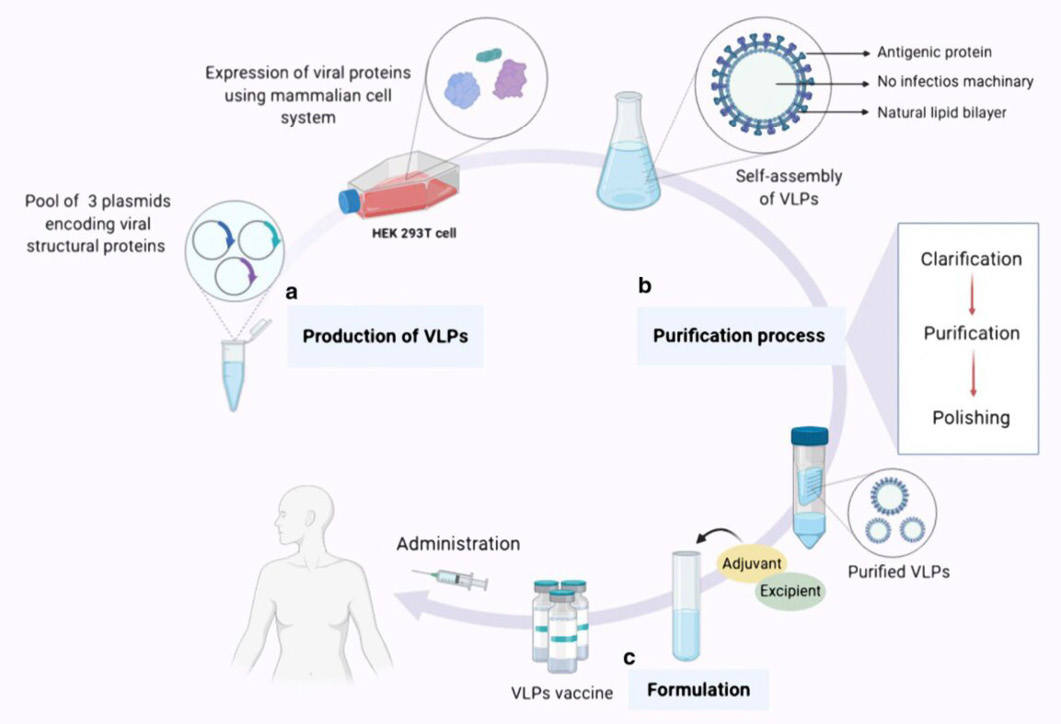
An overview of the expression, purification, and formulation of vaccines based on VLPs. There are typically three steps involved in producing a vaccination based on VLPs. The first step is called “Production,” and it entails cloning the viral structural genes of interest and then expressing the viral proteins that have the potential to self-assemble in an appropriate expression platform (in this case, the HEK293T cell line, a mammalian expression system). The final product of this process is a collection of VLPs in the form of noninfectious particles. b Purification phase, which includes downstream processes such as clarification, purification, and refining, culminates in the acquisition of intact purified VLPs devoid of any residual host detritus. c. The formulation stage, during which adjuvants as well as supplementary ingredients are incorporated into the vaccine formulation to attain a final product that is not only secure but also efficient and effective for vaccination purposes.
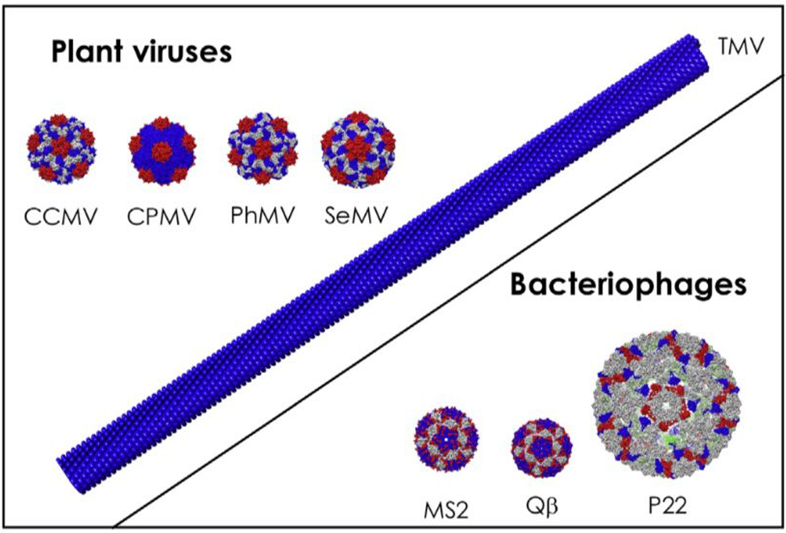
Numerous experimental vaccines have been effectively developed using noninfectious plant VLPs as their foundation. A substrate for antigen expression on the surfaces of over fifty-five distinct plant viruses has been established, which includes but is not limited to the potato X virus (PVX), cucumber mosaic virus (CMV), alfalfa mosaic virus (AIMV), cowpea mosaic virus (CPMV), and papaya mosaic virus (PapMV). TMV, PVX, CPMV, and CCMV are significantly more stable at elevated temperatures as well pH levels, and they are abundantly expressed in native plant hosts.
Formulation of VLP-based vaccines.
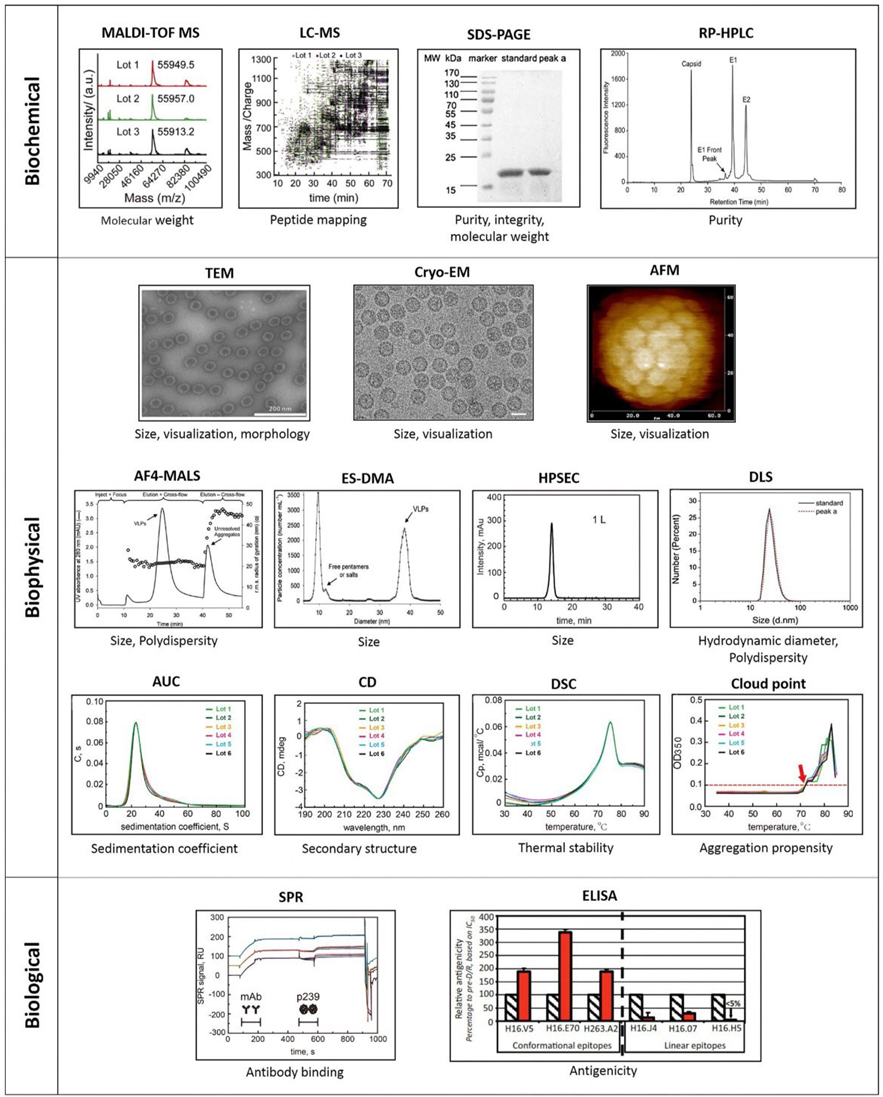
Analytical techniques for characterization of VLPs. A series of analytical tools has been used for biochemical, biophysical and biological characterization of VLPs. Biochemical method: Matrix assisted laser desorption ionization time of flight-mass spectrometry (MALDI-TOF MS), liquid chromatography–mass spectrometry (LC–MS), sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) and reverse phase-high performance liquid chromatography (RP-HPLC). Biophysical method: Transmission electron microscopy (TEM), cryo-electron microscopy (Cryo-EM), atomic force microscopy (AFM), asymmetric flow field-flow fractionation coupled with multiple-angle light scattering (AF4-MALS), electrospray differential mobility analysis (ES-DMA) and high performance size exclusion chromatography (HPSEC), dynamic light scattering (DLS), analytical ultracentrifugation (AUC), Circular dichroism (CD), differential scanning calorimetry (DSC), Cloud point. Biological characterization: Surface plasmon resonance (SRP), enzyme-linked immunosorbent assay (ELISA). MALDI-TOF MS and LC–MS images are reprinted with permission from Elsevier. SDS- PAGE and DLS images are reprinted with permission from Elsevier, RP-HPLC image is reprinted with permission from Elsevier, TEM, A4-MALS and ES-DMA images are reprinted with permission from John Wiley and Sons. Cryo-EM image is reprinted with permission from Elsevier. AFM is reprinted with permission from Elsevier, HPSEC, AUC, CD, DSC, Cloud point and SPR images are reprinted with permission from Elsevier.
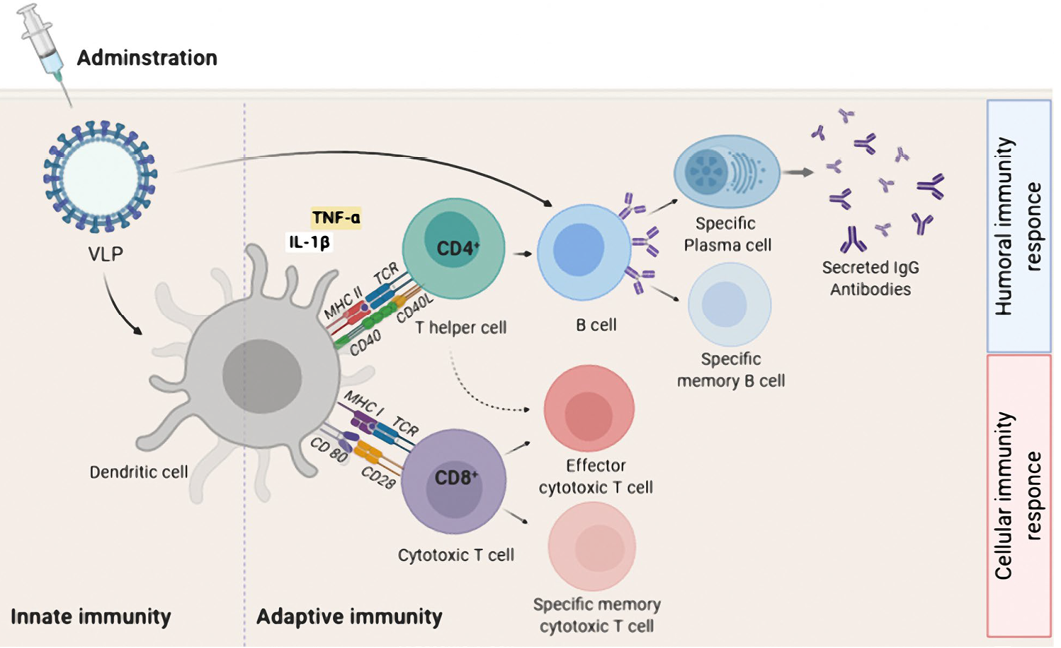
Adaptive immune activation induced by VLP-based vaccine. After administration, a VLP-based vaccine is taken up by APC such as dendritic cells. The phagocytosed VLP-based vaccine is processed and presented by both MHC-II and MHC-I for detection by CD4+ and CD8+T cells, respectively. For induction of humoral immune responses, B cells interact with CD4+ T helper cell (TH) to uptake VLP-based vaccine by B cell receptor. The interaction between CD4+ TH cells and B cells occurs for sufficient secretion of IgG antibodies by plasma cells as well as the generation of B memory cells. For induction of cellular immunity responses, immature CD8+ cytotoxic T lymphocytes (CTL) proliferate and differentiate into effector and specific memory CTL. Effector CD4+ TH cells, increase antigen presenting by APC by secreting cytokines, and also assist activated CTL.
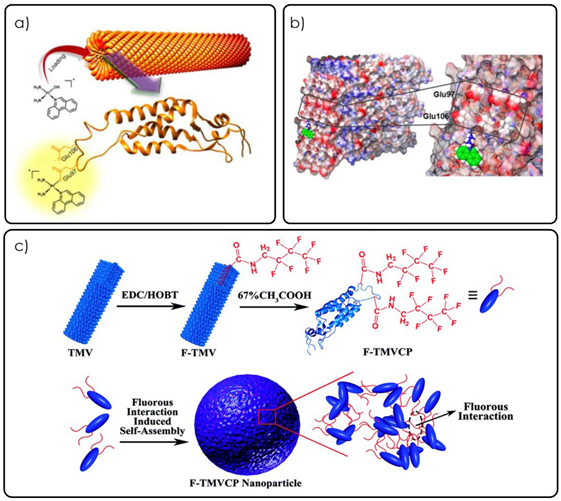
Different mechanisms of loading platinum-based drugs such as phen Pt and cisplatin into TMV a) Graphical abstract from Lippard, S.J., et al. showing the loading schematic for phenPt loading into wild-type TMV particles (reproduced with permission from ref (Vernekar et al., 2018b) Graphical illustration of the phenPt docking onto the Glu97 and Glu106 residues of TMV as discovered through matrix-assisted laser desorption/ionization–mass spectrometry and nuclear magnetic resonance spectroscopy c) Graphical schematic of the assembly of TMV spherical nanoparticles using a fluorous ponytail interaction (F-TMVCP). Cisplatin was loaded into the F-TMVCP using the same glutamic residues as in b (not shown) (reproduced with permission from ref (Gao et al., 2018).
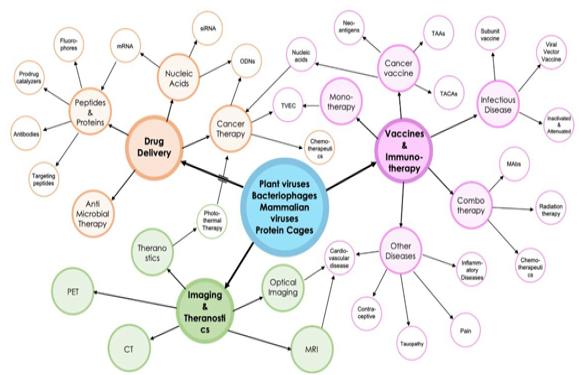
Bubble diagram of the proposed drug delivery and imaging/theranostic platforms discussed in this review by plant viruses, bacteriophages, mammalian viruses, and protein cages.