NS5A Resistance Associated Mutations to Daclatasvir in Hepatitis C Virus Genotype 3a Treatment Naive and SOF/DCV Treatment Failure Patients
NS5A Resistance Associated Mutations to Daclatasvir in Hepatitis C Virus Genotype 3a Treatment Naive and SOF/DCV Treatment Failure Patients
Saima Younas and Aleena Sumrin*
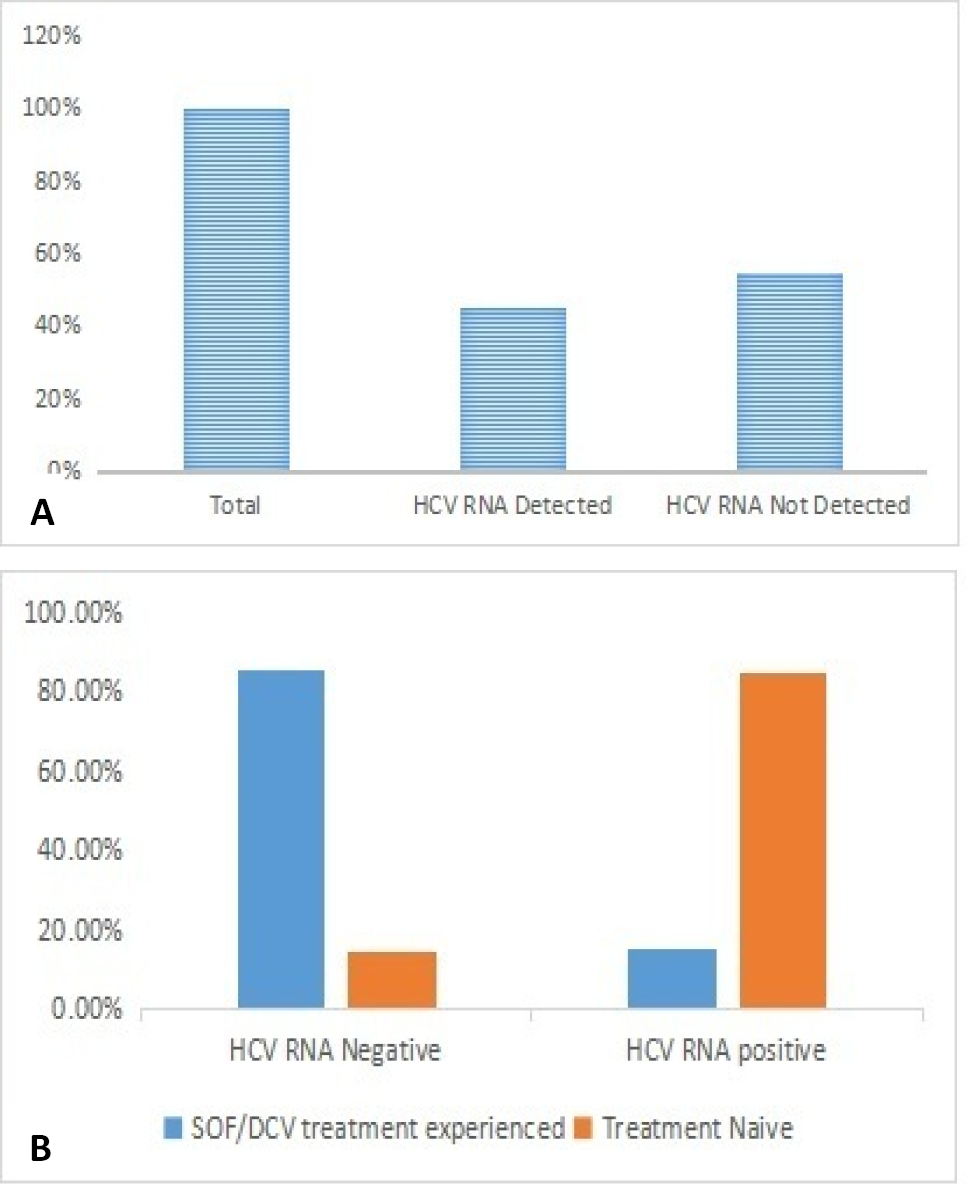
Frequency of HCV RNA detected and not detected (A) and HCV treatment experienced and treatments Naïve (B) patients in study group.
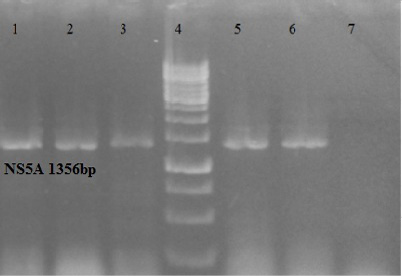
NS5A amplified gene of HCV GT-3a, Lane 1, 2, 3, 5 study samples, Lane 4, 1kb DNA ladder, lane 6 positive control, lane 7 negative control.
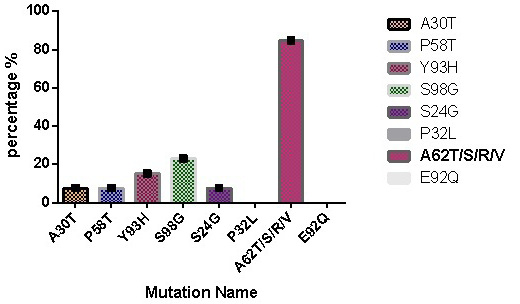
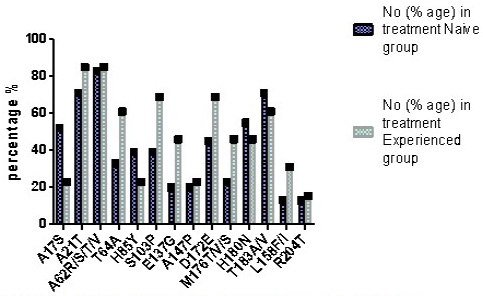
Prevalence of DCV resistance associated amino acid mutations in treatment experienced group.
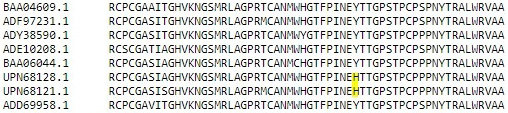
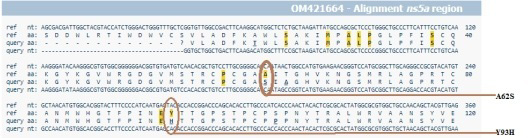
Sequence alignment of reported sequences with Y93H mutation with reference sequence.
HCV NS5A gene sequence alignment using Geno2Pheno (HCV) tool, indicating presence of double amino acids mutations.
Amino acid mutations in treatments naïve and treatment experienced groups.
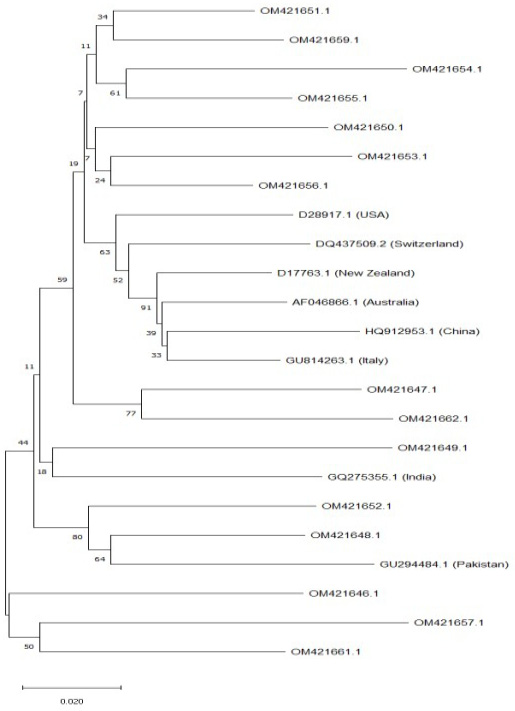
Phylogenetic tree of studied sequences with reference sequences.