Quantitative Studies in Upland Cotton (Gossypium hirsutum L.) using Multivariate Techniques
Muhammad Rizwan1*, Jehanzeb Farooq1, Muhammad Farooq1,2, Aqeel Sarwar4, Abid Ali3, Farrukh Ilahi1, Muhammad Asif1 and Ghulam Sarwar1
1Cotton Research Station, Ayub Agricultural Research Institute, Faisalabad, Pakistan; 2School of Biology and Environmental Science, Science and Engineering Faculty, Queensland University of Technology, Brisbane, Australia; 3Reigonal Agricultural Research Institute, Bahawalpur, Pakistan; 4Crop Sciences Institute, National Agricultural Research Centre, Islamabad, Pakistan.
Abstract | Sustainable production of cotton be contingent upon development of genotypes having better yield, tolerance with respect to abiotic and biotic stresses and enhanced fiber quality. Forty five elite lines of cotton were used to evaluate the genetic variability for fifteen parameters viz., plant height (cms), days taken in 50% flowering, monopodia plant-1, sympodia plant-1, boll weight (g), No. of bolls plant-1, ginning out turn (%), lint index(g), seed index(g), 2.5 percent span length(mm), bundle strength(g/tex), micronaire (µg/in), fibre elongation (%), uniformity ratio and yield plant-1(g). Using Mahalanobis D2 analysis, these parameters were assembled into seven clusters. Among these clusters, cluster I and VII were largest each having nine and eight genotypes respectively, followed by cluster VI having seven genotypes. According to the illustrations by using hierarchical cluster analysis, total genotypes were grouped in seven clusters with 11 genotypes in cluster VI after that cluster II comprising 9 genotypes. The random distribution among genotypes showed that no parallelism exists amongst genetic and geographical diversity. First seven components in principal component analysis (PCA) having eigenvalue more than 1, showed 91.131 % the cumulative variance, while PC-1 alone showed 32.47 % variance. Hierarchical cluster analysis and PCA provided an opportunity to identify subgroups of clusters at different stages, so that every single subgroup may be analyzed critically and it will be helpful for incorporation of desirable characters in future breeding programmes.
Received | September 14, 2020; Accepted | January 11, 2021; Published | February 23, 2021
*Correspondence | Muhammad Rizwan, Cotton Research Station, Ayub Agricultural Research Institute, Faisalabad, Pakistan; Email: muhammadrizwan29@gmail.com
Citation | Rizwan, M., J. Farooq, M. Farooq, A. Sarwar, A. Ali, F. Ilahi, M. Asif and G. Sarwar. 2021. Quantitative studies in upland cotton (Gossypium hirsutum L.) using multivariate techniques. Pakistan Journal of Agricultural Research, 34(1): 113-120.
DOI | http://dx.doi.org/10.17582/journal.pjar/2021/34.1.113.120
Keywords | Multivariate, Analysis, Cotton, Cluster, Diversity
Introduction
Cotton (Gossypium hirsutum L.), generally grown for its fiber in more than eighty countries of the world, is considered chief cash earning commodity and backbone of Pakistan’s economy by contributing 0.8% to GDP and 4.1% to value added in agriculture (Anonymous, 2019). The cotton industry needs higher quantity and quality of raw cotton due to revolution in textile technology. Therefore, it is need of the time to develop high yielding cotton genotypes with superior fiber quality. The development of genotypes having genetically superior qualitative and quantitative traits is inevitable to combat with biotic and abiotic stresses (Bakhtavar et al., 2015). Exploitation of genetic diversity is very useful to identify desirable genotypes in existing germplasm for cotton improvement (Asha et al., 2013). The genetic diversity is assumed as major prerequisite for breeding program to overcome unexpected effects on crop plants due to frequent changes in climatic conditions (Rathinavel, 2017; Jarwar et al., 2019). The selection of genotypes having wider genetic diversity for numerous yield and fiber quality parameters is vital for future strategies of cotton breeding (Shabbir et al., 2016).
Multivariate analysis is being used as major tool to explore genetic divergence in genotypes (Jarwar et al., 2019). Normally, hierarchical cluster analysis, Mahalanobis D2 statistics and principal component analysis are used to explore genetic divergence in multivariate studies. Cluster analysis is important because it is helpful for in-depth analysis by splitting the clusters into sub clusters. The PCA is multivariate tool which extracts most valuable facts from data array into principal components (Sharma, 2006). While partitioning the total variation, PCA is very appropriate statistical tool which is useful to obtain suitable parents for effective breeding strategies (Akter et al., 2009; Nazir et al., 2013). Multivariate analysis approaches on cotton genotypes were accomplished by scientists, which enabled them to categorize the existing germplasm into distinctive clusters based upon fibre quality and yield traits (Shakeel et al., 2015). The experiment aimed to investigate genetic divergence and association amongst forty-five cotton genotypes and categorize them into different classes using multivariate techniques. So, that this information can be utilized in further breeding studies for heterosis and ultimately resulting in improvement of yield and quality parameters by selecting highly divergent parents.
Materials and Methods
Plant materials and location properties
Forty-five cotton strains (Table 1), were studied during Kharif 2019-20 at Cotton Research Station, Faisalabad, Punjab, Pakistan which is situated at altitude of 184 m at 31° 21′ 52′ N 72° 59′ 40′ E having mean annual rainfall 43.2 mm, humidity 61.64 % and temperature 24.25 °C. The trial was sown on 1st May to evaluate the genetic variability of genotypes for morphological traits, fibre parameters and yield.
Management and design of trial
The genotypes were arranged in completely randomized block design having 3 replicates. The plot size of each entry was 4.54 m × 3.03 m, which comprised of four rows having plant x plant spacing 30 cms, while row x row spacing was 75 cms. Delinted seed of each entry was treated with fungicide and insecticide before sowing on beds. Gap filling was practiced after one week of sowing to ensure the plant population. Pre-emergence weedicide was applied before sowing and after germination manual weeding was practiced in the trial. Thinning was done at 25 days after sowing. Recommended dose of fertilizer was used viz., N: P: K @ 80:35:30 kg/ha respectively. Twelve irrigations were applied to the experiment during the season while plant protection measures were adopted as per requirement.
Traits measurement
Ten representative and undamaged plants from each plot were randomly marked for identification and data collection of parameters viz., plant height (cms), days to 50 % flowering, monopodia plant-1, sympodia plant-1, boll weight (g), No. of bolls plant-1, ginning out turn (%), lint index (g), seed index (g), 2.5 percent span length (mm), bundle strength (g/tex), micronaire (µg/in), fibre elongation (%), uniformity ratio and yield plant-1(g). Plant height (cms) was recorded with measuring rod from the base to the tip of the plant. Days to 50 % flowering was obtained by counting the days from sowing date to flower appearance on 50% plants. Monopodia plant-1 and sympodia plant-1 were calculated by counting the number of indirect and direct fruit bearing branches respectively. Boll weight (g) was calculated by picking 50 bolls from top, middle and base of each guarded plant and dividing the total weight by number of bolls. No. of bolls plant-1 was obtained by counting the total no. of bolls of guarded plants. Yield plant-1(g) was obtained on plot basis. The seed of each entry was ginned with single roller ginning machine and lint gained from samples was weighed to calculate GOT % with the formula given below:
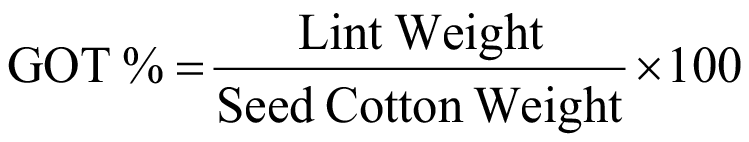
Fiber characteristics
At full maturation, the seed cotton was picked carefully and ginned after drying under sunshine. The fiber quality traits were assessed by Uster-1000 High Volume Instrumentation (HVI) (Sasser, 1981).
Statistical analysis
Tochers’ method was used for Mahalanobis D2 analysis as given by (Rao, 1952). Agglomerative heirarchial clustering method was worked out for cluster analysis following the method demonstrated by (Anderberg, 1993). Principle component analysis (PCA) was performed as given by (Jackson, 1991).
Table 1: List of Genotypes included in the study.
|
Sr. No. |
Name of Genotype |
Status |
Source of Seed |
Sr. No. |
Name of Genotype |
Status |
Source of Seed |
|
1 |
520/18 |
Cultivar |
Cotton Research Station, Faisalabad |
24 |
6030/18 |
Cultivar |
Cotton Research Station, Faisalabad |
|
2 |
521/18 |
Cultivar |
Cotton Research Station, Faisalabad |
25 |
6034/18 |
Cultivar |
Cotton Research Station, Faisalabad |
|
3 |
522/18 |
Cultivar |
Cotton Research Station, Faisalabad |
26 |
6037/18 |
Cultivar |
Cotton Research Station, Faisalabad |
|
4 |
523/18 |
Cultivar |
Cotton Research Station, Faisalabad |
27 |
6039/18 |
Cultivar |
Cotton Research Station, Faisalabad |
|
5 |
524/18 |
Cultivar |
Cotton Research Station, Faisalabad |
28 |
FH-496 |
Advanced Line |
Cotton Research Station, Faisalabad |
|
6 |
494/18 |
Cultivar |
Cotton Research Station, Faisalabad |
29 |
FH-497 |
Advanced Line |
Cotton Research Station, Faisalabad |
|
7 |
495/18 |
Cultivar |
Cotton Research Station, Faisalabad |
30 |
FH-500 |
Advanced Line |
Cotton Research Station, Faisalabad |
|
8 |
455/18 |
Cultivar |
Cotton Research Station, Faisalabad |
31 |
FH-505 |
Advanced Line |
Cotton Research Station, Faisalabad |
|
9 |
505/18 |
Cultivar |
Cotton Research Station, Faisalabad |
32 |
6006/15 |
Cultivar |
Cotton Research Station, Faisalabad |
|
10 |
456/18 |
Cultivar |
Cotton Research Station, Faisalabad |
33 |
FH-419 |
Advanced Line |
Cotton Research Station, Faisalabad |
|
11 |
6035/17 |
Cultivar |
Cotton Research Station, Faisalabad |
34 |
FH-504 |
Advanced Line |
Cotton Research Station, Faisalabad |
|
12 |
452/18 |
Cultivar |
Cotton Research Station, Faisalabad |
35 |
FH-492 |
Advanced Line |
Cotton Research Station, Faisalabad |
|
13 |
458/18 |
Cultivar |
Cotton Research Station, Faisalabad |
36 |
FH-495 |
Advanced Line |
Cotton Research Station, Faisalabad |
|
14 |
6028/17 |
Cultivar |
Cotton Research Station, Faisalabad |
37 |
FH-498 |
Advanced Line |
Cotton Research Station, Faisalabad |
|
15 |
6038/17 |
Cultivar |
Cotton Research Station, Faisalabad |
38 |
FH-453 |
Advanced Line |
Cotton Research Station, Faisalabad |
|
16 |
6061/17 |
Cultivar |
Cotton Research Station, Faisalabad |
39 |
FH-455 |
Advanced Line |
Cotton Research Station, Faisalabad |
|
17 |
6007/18 |
Cultivar |
Cotton Research Station, Faisalabad |
40 |
FH-503 |
Advanced Line |
Cotton Research Station, Faisalabad |
|
18 |
6008/18 |
Cultivar |
Cotton Research Station, Faisalabad |
41 |
FH-506 |
Advanced Line |
Cotton Research Station, Faisalabad |
|
19 |
6009/18 |
Cultivar |
Cotton Research Station, Faisalabad |
42 |
FH-507 |
Advanced Line |
Cotton Research Station, Faisalabad |
|
20 |
6011/18 |
Cultivar |
Cotton Research Station, Faisalabad |
43 |
FH-510 |
Advanced Line |
Cotton Research Station, Faisalabad |
|
21 |
6019/18 |
Cultivar |
Cotton Research Station, Faisalabad |
44 |
FH-511 |
Advanced Line |
Cotton Research Station, Faisalabad |
|
22 |
6021/18 |
Cultivar |
Cotton Research Station, Faisalabad |
45 |
FH-142* |
Advanced Line |
Cotton Research Station, Faisalabad |
|
23 |
6023/18 |
Cultivar |
Cotton Research Station, Faisalabad |
Table 2: Analysis of variance for yield and related parameters in cotton (Gossypium hirsutum L.).
Results and Discussion
Highly significant differences were observed in analysis of variance among 45 genotypes of cotton for 15 quantitative parameters (Table 2). The genetic divergence depicted by 15 parameters was illustrated in Table 3 and Figure 1, which showed the contribution of each trait to the total genetic divergence. All the forty five cotton strains were congregated into seven clusters based upon D2 statistics by using Tocher’s method which depends upon the principle that the inter-cluster D2 values must be higher than intra-cluster D2 average values (Table 4). There was random distribution of forty five strains among seven clusters having maximum strains in cluster I (9 strains) while 8 strains in cluster VII and 7 strains in cluster VI. Six strains were present in cluster III and IV while 5 strains in cluster II and 4 strains in cluster V. Diagrammatic relationship among clusters was illustrated in Table 5, keeping in view the mean of intra and inter cluster D2 values. Similar findings were observed by (Singh et al., 2012; Singh and Dubey, 2011; Srinivasulu et al., 2010; Eswara et al., 2009; Gopinath et al., 2009). Cluster-V showed maximum intra-cluster distance whereas it was minimum for cluster-VI and VII, which indicates that the strains in cluster-V was of diverse genetic makeup and these strains might belong to different genetic pool, whereas the inclinations were contrary for cluster VI and VII (Table 5, Figure 2). The strains of clusters IV and VI showed maximum inter-cluster distance which indicates existence of divergence in genetic makeup of strains among those clusters. Lowest inter-cluster distance was exhibited among strains in cluster-I and II, demonstrating the similarity among strains of this group regarding all parameters. The grouping array of the strains in clusters and inter-cluster distances showed that very less domestication was occurred and ultimately no parallelism amongst geographical distribution and genetic divergence of studied strains. Similar observations were described by (Singh et al., 2012; Mohanty and Prusti, 2002; Mohanty, 2001; Lokhande et al., 1987). While considering inter- cluster distances among the groups, best desirable segregants may be obtained by crossing the strains of clusters IV and VI after confirming their general combining ability. These outcomes are in line with the findings of (Asha et al., 2013).
According to hierarchial clustering (Ward’s minimum variance) method, forty five strains were assembled into 7 clusters (Table 6). Cluster-VI was biggest comprising 11 strains after that cluster-II having 9 strains, cluster-V having 7 strains, cluster-I having 6 strains, cluster-IV and-VII (5 strains each) and cluster-III (2 strains). Average intra and inter-cluster Euclidean distance (D2) was computed following
Table 3: Contribution of parameters studied towards genetic variability in 45 strains of cotton (Gossypium hirsutum L.).
|
Source |
Times ranked first |
Contribution % |
|
Plant height (cm) |
39 |
4.89 |
|
Days to 50% flowering |
66 |
7.64 |
|
No. of monopodia plant-1 |
165 |
22.32 |
|
No. of Sympodia plant-1 |
3 |
0.38 |
|
Boll weight (gm) |
44 |
6.5 |
|
No. of Bolls plant-1 |
22 |
3.76 |
|
Ginning out-turn % |
4 |
0.42 |
|
Lint index(g) |
26 |
4.52 |
|
Seed index(g) |
193 |
23.49 |
|
2.5 % Span length(mm) |
31 |
3.22 |
|
Bundle strength (g/tex) |
55 |
5.24 |
|
Microniare (µg/in) |
45 |
4.58 |
|
Fibre Elongation % |
4 |
0.49 |
|
Uniformity ratio |
20 |
2.87 |
|
Seed cotton Yield plant-1 |
63 |
9.66 |
Ward’s minimum variance technique and was illustrated in Table 7. Cluster-II exhibited maximum intra-cluster Euclidean2 distance having value of 259.54 after that clusters-I (181.65), III (151.93), IV (149.78), VII (115.33), VI (107.84) and V (0.00) demonstrating maximum variability contained by cluster-II related to other clusters. The range of inter-cluster Euclidean distance (D2) was from 169.13 (among clusters VI and VII) to 769.72 (clusters VI and V). According to the study, the clusters VI and VII are highly divergent, consequently, the strains from these two clusters may further be exploited in future breeding strategies for heterosis studies. Similar conclusions were submitted by (Asha et al., 2013; Lakshmi et al., 2009; Srinivasulu et al., 2010; Altaher and Singh, 2003).
Principal Component Analysis (PCA) revealed that Eigen values of first seven components was more
Table 4: Distribution of 45 cotton strains in various clusters using Tocher’s method.
|
Cluster No. |
Name of genotype(s) |
No. of genotypes |
|
I |
FH-498, FH-453, 6038/17, 522/18, 6021/18, FH-507, 6035/17, FH-505, 6011/18 |
9 |
|
II |
523/18, 6034/18, FH-496, FH-452, FH-142 |
5 |
|
III |
6030/18, 524/18, 494/18, 6007/18, FH-510, 520/18 |
6 |
|
IV |
FH-458, 6028/17, 6061/17, FH-511, 6023/18, FH-419 |
6 |
|
V |
6008/18, FH-503, FH-504, FH-495 |
4 |
|
VI |
6019/18, 521/18, 455/18, 505/18, 456/18, 6006/15, FH-492 |
7 |
|
VII |
495/18, FH-455, 6009/18, 6037/18, 6039/18, FH-497, FH-500, FH-506 |
8 |
Table 5: Average intra- (Diagonal values) and inter (Above diagonal values) cluster divergence D2 and D* values of among 7 clusters of 45 Cotton (Gossypium hirsutum L.) genotypes.
|
Clusters |
I |
II |
III |
IV |
V |
VI |
VII |
|
I |
25.21 (5.02) |
41.37(6.43) |
76.41(8.74) |
61.27(7.83) |
82.84(9.10) |
231.95(15.23) |
173.59(13.18) |
|
II |
31.42(5.60) |
59.65(7.72) |
68.93(8.30) |
81.33(9.02) |
183.29(13.54) |
128.36(11.33) |
|
|
III |
48.96(6.99) |
79.86(8.94) |
105.2(10.26) |
123.81(11.13) |
104.96(10.25) |
||
|
IV |
51.75(7.19) |
140.68(11.86) |
241.47(15.54) |
155.65(12.48) |
|||
|
V |
83.62(9.14) |
192.58(13.88) |
152.81(12.36) |
||||
|
VI |
0.00(0.00) |
88.61(9.41) |
|||||
|
VII |
0.00(0.00) |
* Values in parentheses are D values.
Table 6: Distribution of 45 cotton strains in various clusters using Ward’s minimum variance method.
|
Cluster no |
Name of genotype(s) |
No. of genotypes |
|
I |
455/18, 6008/18, 456/18, FH-497, FH-500, FH-507 |
6 |
|
II |
520/18, 495/18, 6028/17, 6038/17, 505/18, FH-495, FH-503, FH-506, FH-142 |
9 |
|
III |
522/18, FH-455 |
2 |
|
IV |
6061/17, 6007/18, FH-419, FH-453, FH-511 |
5 |
|
V |
494/18, FH-505, 6035/17, 6021/18, 6034/18, FH-496, FH-492 |
7 |
|
VI |
521/18, 523/18, 524/18, FH-452, 6009/18, 6011/18, 6023/18, 6030/18, 6037/18, 6006/15, FH-504 |
11 |
|
VII |
FH-458, 6019/18, 6039/18, FH-498, FH-510 |
5 |
Table 7: Average Eucledian2 (intra and inter-cluster) values among clusters of 45 Cotton (Gossypium hirsutum L.) strains.
|
Cluster No. |
I |
II |
III |
IV |
V |
VI |
VII |
|
I |
181.65 |
407.91 |
275.83 |
272.82 |
621.95 |
277.34 |
256.86 |
|
II |
|
259.54 |
293.51 |
459.16 |
712.65 |
556.16 |
553.7 |
|
III |
|
|
151.93 |
279.3 |
631.29 |
230.42 |
254.59 |
|
IV |
|
|
|
149.78 |
378.43 |
282.42 |
196.98 |
|
V |
|
|
|
|
0 |
769.72 |
536.83 |
|
VI |
|
|
|
|
|
107.84 |
169.13 |
|
VII |
|
|
|
|
|
|
115.33 |
Table 8: Factor loadings and Eigen values of principal components.
|
PC1 |
PC2 |
PC3 |
PC4 |
PC5 |
PC6 |
PC7 |
|
|
Eigen value (Root) |
3.246 |
2.874 |
2.024 |
1.628 |
1.367 |
1.0354 |
1.024 |
|
% Var. Exp. |
32.472 |
17.654 |
11.107 |
9.421 |
8.223 |
6.015 |
6.239 |
|
Cum. Var. Exp. |
32.472 |
50.126 |
61.233 |
70.654 |
78.877 |
84.892 |
91.131 |
than one and contributed 91.131 percent to the total variability (Table 8). Therefore, inference can be drawn that the most valuable information of data set was present in first seven principal components. Similar observations were previously described by (Kumari et al., 2019; Shah et al., 2018; Latif et al., 2015; Kaleri et al., 2015; Saeed et al., 2014).
Conclusions and Recommendations
The PCA and hierarchical cluster analysis confirmed findings of each other. Non-correspondence between geographic diversity and genetic divergence was confirmed by adopting these three methods of grouping. Hierarchical cluster analysis provided an opportunity to identify subgroups of clusters at different stages, so that every single subgroup may be analyzed critically and will be helpful for incorporation of desirable characters in future breeding programs.
Novelty Statement
Hierarchical cluster analysis and principal component analysis provided an opportunity to identify subgroups of clusters at different stages, so that every single subgroup may be analyzed critically and it will be helpful for incorporation of desirable characters in future breeding programmes.
Author’s Contribution
Muhammad Rizwan: Performed the experiment and collected data.
Jehanzeb Farooq: Managed overall crop and gave technical input.
Muhammad Farooq: Helped in paper write up,
Muhammad Aqeel Sarwar: Performed the statistical analysis.
Farrukh Ilahi: Helped in data collection.
Abid Ali: Helped in data analysis.
Muhammad Asif: collected the literature.
Ghulam Sarwar: Supervised the study.
Conflict of interest
The authors have declared no conflict of interest.
References
Akter, A., M.J. Hasan, A.K. Paul, M.M. Mutlib and M.K. Hossain. 2009. Selection of parent for improvement of restorer line in rice (Oryza sativa L.). SAARC. J. Agric., 7: 43-50.
Altaher, A.F. and R.P. Singh. 2003. Genetic diversity studies in upland cotton (Gossypium hirsutum L.). J. Ind. Soc. Cotton Improv., 28: 158-163.
Anderberg, M.R., 1993. Cluster analysis for application. Academic Press, New York.
Anonymous, 2019. Pakistan economic survey, Govt. Pak., Fin. Div. Adv. Wing, Islamabad.
Asha, R., M.L. Ahamed, D.R. Babu and P.A. Kumar. 2013. Multivariate Analysis in Upland Cotton (Gossypium hirsutum L.). Madras Agric. J., 100(4-6): 333-335.
Bakhtavar, M.A., I. Afzal, S.M.A. Basra, A.U.H. Ahmad and M.A. Noor. 2015. Physiological strategies to improve the performance of Spring Maize (Zea mays L.) planted under early and optimum sowing conditions. PLoS One, 10(4): 1-15. https://doi.org/10.1371/journal.pone.0124441
Eswara, R.G., V.C. Reddy, M.L. Ahamed, V.S. Rao, C.P. Rao and V.B. Reddy. 2009. Genetic divergence in upland cotton (Gossypium hirsutum L.). Andhra Agric. J., 56: 309-314.
Gopinath, M., S. Rajamani, R.K. Naik and C.M. Rao. 2009. Genetic divergence for lint characters in upland cotton (Gossypium hirsutum L.). J. Cotton Res. Dev., 23: 46-48.
Jackson, J.E., 1991. A user’s guide to principal components. John Wiley and Sons Inc., New York. https://doi.org/10.1002/0471725331
Jarwar, A.H., X. Wang, M.S. Iqbal, Z. Sarfraz, L. Wang, Q. Ma and F. Shuli. 2019. Genetic divergence on the basis of principal component, correlation and cluster analysis of yield and quality traits in cotton cultivars. Pak. J. Bot., 51(3): 1143-1148. https://doi.org/10.30848/PJB2019-3(38)
Kaleri, A.A., S.Y. Rajput, G.A. Kaleri and J.A. Marri. 2015. Analysis of Genetic diversity in genetically modified and non-modified cotton (Gossypium hirsutum L.) genotypes. IOSR J. Agric. Vet. Sci., 8(12): 70-76.
Kumari V.N. and P. Gunasekaran. 2019. Evaluation of genetic diversity in cotton (Gossypium barbadense L.) germplasm for yield and fibre attributes by principle component analysis. Int. J. Curr. Microbiol. App. Sci., 8(04): 2614-2621. https://doi.org/10.20546/ijcmas.2019.804.304
Lakshmi, G.V., V.C. Reddy, C.P. Rao, J.S. Babu and R. Srinivasulu. 2009. Multivariate analysis of genetic diversity in upland cotton (Gossypium hirsutum L.). Andhra Agric. J., 56: 8-15.
Latif, A., M. Bilal, S.B. Hussain and F. Ahmad. 2105. Estimation of genetic divergence, association, direct and indirect effects of yield with other attributes in cotton (Gossypium hitsutum L.) using biplot correlation in and path coefficient analysis. J. Trop. Pl. Res., 2(2): 120-126.
Lokhande, G.D., B.B. Pawar, A.D. Dumbre and R.Y. Thete. 1987. Genetic divergence in garlic (Allium sativum L.). Curr. Res. Rep., 3: 98-99.
Mohanty, B.K. and A.M. Prusti. 2002. Mahalanobis’s generalized distance analysis in onion. J. Res. Crop., 3: 142-144.
Mohanty, B.K., 2001. Analysis of genetic divergence in Kharif onion. In. J. Hortic., 58: 260-263.
Nazir, A., J. Farooq, A. Mahmood, M. Shahid and M. Riaz. 2013. Estimation of genetic diversity for CLCuV, earliness and fiber quality traits using various statistical procedures in different crosses of Gossypium hirsutum L. Vestnik. Orel. Gau., 43: 2-9.
Rao, C.R., 1952. Advanced statistical methods in biometrical research. John Wiley and Sons Inc. New York. pp. 236-272.
Rathinavel, K., 2017. Exploration of genetic diversity for qualitative traits among the extant upland cotton (Gossypium hirsutum L.) varieties and parental lines. Int. J. Curr. Microbiol. App. Sci., 6(8): 2407-2421. https://doi.org/10.20546/ijcmas.2017.608.285
Saeed, F., J. Farooq, A. Mahmood, M. Riaz, T. Hussain and A. Majeed. 2014. Assessment of genetic diversity for cotton leaf curl virus (CLCuD), fibre quality and some morphological traits using different statistical procedures in Gossypium hirsutum L. Aust. J. Crop Sci., 8(3): 442- 447.
Sasser, P.E., 1981. The basics of high volume instrumentation for fiber testing. Proc. Beltwide Cotton Prod. Res. Conf., New Orleans, LA 4-8 Jan., Natl. Cotton Counc. Am., Memphis, TN. pp. 191-193.
Shabbir, R.H., Q.A. Bashir, A. Shakeel, M.M. Khan, J. Farooq, S. Fiaz, B. Ijaz and M.A. Noor. 2016. Genetic divergence assessment in upland cotton (Gossypium Hirsutum L.) Using various statistical tools. J. Glob. Innov. Agric. Soc. Sci., 4(2): 62-69. https://doi.org/10.22194/JGIASS/4.2.744
Shah, A.S., J.S., Khan, K. Ullah and O.U. Sayal. 2018. Genetic diversity in cotton germplasm using multivariate analysis. Sarhad J. Agric., 34(1): 130-135. https://doi.org/10.17582/journal.sja/2018/34.1.130.135
Shakeel, A., I. Talib, M. Rashid, A. Saeed, K. Ziaf and M.F. Saleem. 2015. Genetic diversity among upland cotton genotypes for quality and yield related traits. Pak. J. Agric. Sci., 52(1): 73-77.
Sharma, J.R., 2006. Statistical and biometrical techniques in plant breeding. New Age International Publishers. pp. 432.
Singh, R.K. and B.K. Dubey. 2011. Studies on genetic divergence in onion advance lines. Ind. J. Hortic., 68(1): 123-127.
Singh, R.K., B.K. Dubey and R.P. Gupta. 2012. Studies on variability and genetic divergence in garlic (Allium sativum L). J. Spices Arom. Crop, 21(2): 136-144.
Srinivasulu, P., J.S.V. Samba Murthy, P.V.R. Kumar and V.S. Rao. 2010. Multivariate analysis of genetic diversity in upland cotton (Gossypium hirsutum L.). Andhra Agric. J., 57: 148-155.
To share on other social networks, click on any share button. What are these?







