Optimization of Tannase Production from Raoultella ornithinolytica using Corn (Zea mays) Leaves in Solid State Fermentation
Optimization of Tannase Production from Raoultella ornithinolytica using Corn (Zea mays) Leaves in Solid State Fermentation
Hafiz Abdullah Shakir1*, Iqra Javed1, Muhammad Irfan2*, Shaukat Ali3, Muhammad Khan1, Farah Rauf Shakoori1, Javed Iqbal Qazi1 and
Muhammad Abrar Yousaf1
1Department of Zoology, University of the Punjab, Lahore, Pakistan
2Department of Biotechnology, University of Sargodha, Sargodha, Pakistan
3Department of Zoology, Government College University, Lahore, Pakistan
ABSTRACT
Tannase has significant importance due to its various industrial applications. Tannase production using pure tannic acid as substrate is very expensive especially at industrial level. In present study, various physical parameters and medium components were optimized for maximum tannase production employing Raoultella ornithinolytica in solid state fermentation (SSF) using corn (Zea mays) leaves as substrate to reduce its production cost. The maximum tannase production was obtained with 60% initial substrate moisture contents, tap water as enzyme extraction medium with 2 mL volume, 45°C incubation temperature, pH 7, 300 µL inoculum size, 24 h incubation period in agitated condition with substrate particle size of 4mm during one factor at a time optimization. Concentrations of medium components (3.75% tannic acid, 0.75% K2HPO4 and 1.25% yeast extract) were optimized with central composite design of response surface methodology. Tannase characterization data revealed that 5.0 pH, 30°C temperature, 60 minutes incubation and 0.45% of substrate concentration showed highest tannase activity. The results depict utilization potential of low cost substrate (corn leaves) to reduce the production cost of tannase.
Article Information
Received 10 December 2019
Revised 03 March 2020
Accepted 07 July 2020
Available online 15 July 2022
(early access)
Published 27 February 2023
Authors’ Contribution
HAS conceived the idea and planned the experiments. IJ performed experiments and wrote first draft. MI performed statistical analyses. SA and MK provided technical assistance. FRS proofread the manuscript for English Editing. JIQ technically revised the manuscript. MAY performed experiments.
Key words
Central composite design (CCD), Raoultella ornithinolytica, Response surface methodology (RSM), Solid-state fermentation (SSF), Tannase
DOI: https://dx.doi.org/10.17582/journal.pjz/20191210061234
* Corresponding author: [email protected], [email protected]
0030-9923/2023/0003-1131 $ 9.00/0
Copyright 2023 by the authors. Licensee Zoological Society of Pakistan.
This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
INTRODUCTION
Tannin is naturally occurring polyphenolic compound and 4th most abundant component of plants after cellulose, hemicellulose and lignin (Lokeswari and Kumar, 2013). Tannin acyl hydrolase EC 3.1.1.20 generally known as tannase, is an extracellular enzyme that catalyzes tannin by hydrolyzing its depside and ester bonds and liberates glucose as well as gallic acid (Beena et al., 2011). This inducible enzyme is produced by several microorganisms (Böer et al., 2009; Sharma and John, 2011). Tannase has significant importance from industrial perspective. Its commercial worth is being enhanced in latest years because of its various industrial potential applications (Govindarajan et al., 2016). It has applications in pharmaceutical, food, beverages, cosmetic products, animal feed, leather and chemical production (Aguilar et al., 2007). Production of tannase using pure tannic acid as substrate is very expensive especially at industrial level (Lokeswari and Kumar, 2013). In recent times, interest towards the usage of agricultural residues for tannase production as substrate has remarkably been increased due to their low cost. In Pakistan, the annual production of corn and other crops are in million tons and a significant amount of tannin is present in plant parts like leaves and stem. Different agricultural wastes like palm kernel cake, tamarind seed powder, (Sabu et al., 2005), rice straw powder, sugarcane bagasse (Paranthaman et al., 2010), coffee husk (Battestin and Macedo, 2007), tea stalks (Xiao et al., 2015), cashew bagasse (Liu et al., 2016) and olive mill waste (Aissam et al., 2005) have been described for tannase production in literature.
Tannase can be produced by solid state fermentation (SSF) (Madeira et al., 2011; Wang et al., 2013; Chávez-González et al., 2014) as well as submerged fermentation (Selwal et al., 2010; Böer et al., 2011; Chávez-González et al., 2014). SSF (microorganisms grown over solid substrate where free water is absent or nearly absent) using agricultural wastes is often preferred over submerged liquid fermentation because of cheaper raw material, higher enzymatic quality and activity, lower energy cost, minor water consumption and easier process (Barrios-González, 2012; Lessa et al., 2018; Ferraz et al., 2020).
For optimization of independent variables/parameters for enhanced production of enzyme, both one factor at a time (OFAT) as well as response surface methodology (RSM) approaches are applicable. In OFAT, independent variables are optimized by varying levels of one variable/parameter at a time while all other variables are kept constant. This approach is simple in application and determines the factors affecting enzyme yield (Singh et al., 2011). While RSM is used to design experiments, find optimum independent variable interacting with all other variables and its effect on the response i.e., enzyme production (Khuri and Mukhopadhyay, 2010; de Brito et al., 2017; Tripathi and Lakshmi, 2018). The current study was planned to optimize several physical parameters and medium components with OFAT as well as RSM approaches for maximal tannase production in SSF by using corn (Zea mays) leaves as a substrate. Furthermore, influence of different physical parameters on tannase activity was also measured.
MATERIALS AND METHODS
Screening of tannic acid utilizing potential bacteria
In present study, ten (10) bacterial strains formerly isolated from gut contents of fish were revived on nutrient agar medium and then screened to study their tannase producing potential following Osawa and Walsh (1993). Briefly, ten bacterial strains were streaked (a line) on tannic acid medium (0.5% tannic acid incorporated in 2.8% nutrient agar) and greenish zones were observed after incubation at 37ºC for 24 h. All zone forming potential strains were selected for tannase assay.
Production of crude tannase
For tannase production, 1% inoculum of each zone producing strain was inoculated in production broth i.e., 0.5% tannic acid; 0.275% yeast extract; 0.1% CaCl2 according to Javed (2016). After 24 h incubation at 37°C, each culture was centrifuged at 8000 rpm at 4°C for 15 minutes. Crude enzyme (supernatant) was further used in the assay of tannase enzyme.
Tannase assay
Tannase assay was performed following Miller (1959) using tannic acid (0.5%) in acetate buffer (0.1 M) with pH 5 as substrate and glucose as standard. Crude tannase enzyme (0.5 mL) was added in substrate (0.5 mL) followed by 30 min incubation at 37°C. Then 3 mL di-nitro-salicylic acid was added in solution and boiled for 15 minutes in water bath followed by dilution with 10 mL distilled water. Absorbance was noted at 540 nm against blank by UV-visible spectrophotometer. The amount of tannase that can utilize 1 mM tannic acid substrate in one minute under standard conditions of assay was considered enzyme unit. Bacterial strain showing the highest enzyme production was selected for optimizing the condition for enhanced enzyme synthesis.
Preparation of corn substrate
Corn (Zea mays) leaves were cut into small pieces, dried and properly ground to powder to be used as substrate.
Evaluation of optimal physical parameters
Various physical parameters such as substrate moisture content (50, 60, …, 90%), enzyme extraction mediums (distilled water, tap water, 1% NaCl, acetate buffer of pH 4 and 5, phosphate buffer of pH 6.0 and 7.0, tris-HCl buffer of pH 8 and 9 and glycine NaOH buffer of pH 10 and 11) and their volumes (1, 2, ….6 mL), incubation temperatures (37, 40 and 45°C), production medium pH (3, 5, 7, 9 and 11), inoculum size (1, 2 and 3%), incubation periods (24, 48, 72 and 96 h), agitation effect (shaking at 150 rpm and non-shaking condition), substrate particles size (2.8, 3.4, 4.0 mm) and centrifugation effect were optimized by OFAT method to enhanced tannase production in SSF. Each parameter after optimization was added in further experiments.
Evaluation of optimal medium components
Different salts 0.1% (KCl, NaCl, K2HPO4, KH2PO4, CaCl2, MgSO4), tannic acid concentrations (1.5, 2, 2.5, …, 4.0%) and nitrogen source 0.275% (malt extract, yeast extract and peptone) were optimized to find out the optimal medium components for the maximal tannase production by OFAT method in SSF. Each medium component after optimization was added in further experiments.
Evaluation of optimal concentration of medium components
Optimization of concentration of medium components was conducted by central composite design (CCD) of RSM. Seventeen experiments were performed with three medium components and five level face-centered cube design (-2. -1, 0, +1, +2) of independent variables (Table I) with 17 experiments, while full experimental plan is described in Table III.
The significance of model and regression coefficients was analyzed statistically by analysis of variance (ANOVA). Regression analysis was performed for response (tannase) prediction using second order polynomial equation.
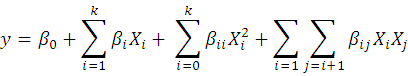
Here y is predicted tannase activity (U/mL), XiXj are independent variables while k is number of applied variables. While βo, βij and βii are the intercept coefficient, interaction coefficient and quadratic coefficient respectively. Data was analyzed using STATISTICA (99th edition) software.
Table I. Levels of medium components used in CCD.
|
Independent variables |
Code |
Levels |
||||
|
-2 |
-1 |
0 |
+1 |
+2 |
||
|
Tannic acid (%) |
A |
3.00 |
3.25 |
3.50 |
3.75 |
4.00 |
|
K2HPO4 (%) |
B |
0.10 |
0.25 |
0.50 |
0.75 |
1.00 |
|
Yeast extract (%) |
C |
0.50 |
0.75 |
1.00 |
1.25 |
1.50 |
Table II. Screening of bacterial strains previously isolated from fish gut content by zone and tannase assay.
|
Bacterial strains |
Zone results |
Tannase (U/ml) |
|
Aeromonas allosaccharophila |
_ |
- |
|
Aeromonas bestiarum |
_ |
- |
|
Aeromonas hydrophila |
_ |
- |
|
Aeromonas media |
_ |
- |
|
Bacillus amyloliquefaciens |
+ |
2.73b±0.09 |
|
Bacillus flexus |
_ |
- |
|
Bacillus pumilus |
_ |
- |
|
Enterobacter aerogenes |
+ |
1.59c±0.08 |
|
Klebsiella oxytoca |
+ |
2.68b±0.06 |
|
Raoultella ornithinolytica |
+ |
3.31a±0.008 |
Mean±SD in column with different letters are significantly different (Tukey’s test, P<0.001).
Evaluation of tannase activity under various parameters
Effect of pH
To determine the effect of various pH on tannase activity, crude enzyme was reacted with substrate prepared in acetate buffer (pH 4 and 5), phosphate buffer (pH 6 and 7), tris-HCl buffer (pH 8 and 9) and glycine NaOH buffer (pH 10 and 11) of 0.1 M and enzyme assay was proceeded as described earlier.
Effect of temperature
The tannase activity was evaluated by incubating the reaction mixture at different temperatures (20, 30, 40…,90°C) in tannase assay. The temperature related to maximum enzyme activity was considered as optimum.
Effect of incubation time
Tannase was incubated at optimum temperature and pH for various times (15, 30, 45…, 75 minutes) in the assay. As the result, incubation time giving the best tannase activity was taken as optimum.
Effect of substrate concentration
To evaluate the substrate concentrations effect, multiple concentrations (0.25, 0.30, 0.35…, 0.60%) of tannic acid were used with all optimized conditions in the assay. Optimum concentration was determined considering maximum enzyme activity.
Statistical analysis
All experiments were conducted in triplicates. Results were presented in mean with standard deviation. Significance and accuracy of the results was analyzed applying Student’s t-test and one-way ANOVA followed by Tukey’s Post Hoc (P<0.05) using computer software IBM SPSS Statistics 20.
Table III. Tannase production by R. ornithinolytica in different concentrations of medium components in experimental statistical CCD design.
|
Run |
Concentration of medium |
Tannase (U/ml) |
Residue value |
|||
|
A (%) |
B (%) |
C (%) |
Observed |
Predicted |
||
|
1 |
3.50 |
0.10 |
1.00 |
121.52 |
101.44 |
20.08 |
|
2 |
3.50 |
0.50 |
0.50 |
125.91 |
129.93 |
-4.03 |
|
3 |
3.75 |
0.75 |
1.25 |
157.04 |
164.96 |
-7.92 |
|
4 |
3.50 |
1.00 |
1.00 |
142.79 |
138.40 |
4.39 |
|
5 |
4.00 |
0.50 |
1.00 |
140.45 |
140.12 |
0.34 |
|
6 |
3.75 |
0.25 |
1.25 |
113.96 |
127.66 |
-13.69 |
|
7 |
3.50 |
0.50 |
1.00 |
112.42 |
110.21 |
2.21 |
|
8 |
3.00 |
0.50 |
1.00 |
144.53 |
127.62 |
16.91 |
|
9 |
3.25 |
0.75 |
0.75 |
154.93 |
158.48 |
-3.56 |
|
10 |
3.25 |
0.25 |
0.75 |
149.22 |
158.55 |
-9.33 |
|
11 |
3.50 |
0.50 |
1.50 |
114.30 |
93.03 |
21.28 |
|
12 |
3.75 |
0.75 |
0.75 |
97.75 |
92.96 |
4.79 |
|
13 |
3.25 |
0.75 |
1.25 |
80.03 |
96.18 |
-16.15 |
|
14 |
3.25 |
0.25 |
1.25 |
27.58 |
49.63 |
-22.05 |
|
15 |
3.75 |
0.25 |
0.75 |
101.15 |
102.26 |
-1.10 |
|
16 |
3.50 |
0.50 |
1.00 |
113.47 |
110.21 |
3.26 |
|
17 |
3.50 |
0.50 |
1.00 |
114.76 |
110.21 |
4.55 |
A, tannic acid; B, K2PHO4; C, yeast extract.
RESULTS
Tannic acid utilizing potential bacterial isolates
In present study, the four bacterial strains out of ten showed greenish clearance zone on tannic acid based medium. During tannase assay, these four strains also showed highly significant (P<0.001) tannase production. The highest tannase production up to 3.31 U/mL was observed by Raoultella ornithinolytica among the strains (Table II).
Optimal physical parameters of tannase
Effect of substrate moisture content
For evaluation of optimum moisture content in corn substrate for tannase production, experiments were carried out with moisture ranging from 50% to 90%. Results depicted that initially enzyme production increased up to 60% then decline was observed. R. ornithinolytica showed the highest tannase production (50.53±0.91 U/mL) with moisture content of 60% in substrate (Fig. 1).
Effect of extraction media
Various solvents i.e., distilled water, tap water, 1% NaCl, acetate buffer of pH 4 and 5, phosphate buffer of pH 6 and 7, tris-HCl buffer of pH 8 and 9 and glycine NaOH buffer of pH 10.0 and 11.0) were used as extraction medium. Results indicated that with tap water maximum tannase (65.73±0.33 U/mL) was produced as shown in Figure 2.
Effect of extraction medium volume
When the volume of extraction medium was changed form 1 mL to 6 mL, the optimal enzyme production (90.41±0.16 U/mL) was noted at 2 mL of optimum extraction medium (tap water) (Fig. 3). With the increase in volume, the enzyme production decreased gradually.
Effect of incubation temperature
Temperature of incubation is also important to influence the bacterial growth. Experiments were performed at different temperatures (37°C, 40°C and 45°C). Results showed the maximum tannase production i.e., 90.57±0.11 U/mL after 24 h incubation at 45°C. While at 30°C and 35°C enzyme units were 71.56±6.76 U/mL and 65.1±1.32 U/mL, respectively (Fig. 4).
Effect of initial medium pH
To determine the optimal pH for enhanced tannase production, different pH ranges were applied from 3.0 to 11.0. Post to 24 h incubation, the highest tannase synthesis was observed at 7 pH with 90.80±0.92 U/mL enzyme value (Fig. 4). First enzyme value increased up to 7.0 pH, then decline in tannase production was detected.
Effect of inoculum size
The maximum enzyme production (91.10±1.03 U/mL) was recorded with 3%. While on 1% and 2%, enzyme values were 56.64±0.75 U/mL and 61.55±0.69 U/mL respectively indicating that with inoculum size enzyme production increased (Fig. 4).
Effect of incubation period
Results at different incubation period (24, 48, 72 and 96 h) were recorded. For R. ornithinolytica, after 24 h the enzyme production (93.33±4.78 U/mL) was maximum (Fig. 4). With further rise in period of incubation, the enzyme value was observed to be decreased.
Effect of agitation
Agitation had significant effect on tannase production. With agitated condition (150 rpm) the enzyme value (96.05±1.39 U/mL) was higher significantly than static condition (61.26±1.76 U/mL) (Fig. 4).
Effect of substrate particle size
Figure 5 indicated the impact of different particle sizes of substrate on tannase production in SSF. Highest enzyme value (121.56±1.96 U/mL) was obtained with large-sized particle (4.0 mm). Minimum enzyme value (87.85±1.33 U/mL) was observed with medium-sized particles (3.4 mm).
Effect of centrifugation
Results exhibited that centrifugation had negative effect on tannase production in SSF. Production was significantly low when centrifugation (103.06±1.39 U/mL) was done than non-centrifuging condition (121.56±1.96 U/mL) (Fig. 6).
Evaluation of optimal medium components
Effect of various salts
Various salts i.e., NaCl, KCl, KH2PO4, K2HPO4, MgSO4 and CaCl2 were used in fermentation medium to check their effect on tannase production. Results indicated highest enzyme production (122.12±1.04 U/mL) with K2HPO4, while lowest tannase value (107.49±2.12 U/mL) was obtained in the presence of KH2PO4 (Fig. 7).
Effect of tannic acid concentrations
Several tannic acid concentrations (1.5%, 2%, 2.5%, …, 4%) were added in medium to check their effect on the tannase production. Highest tannase synthesis (123.24±1.78 U/mL) was obtained at 3.5% of tannic acid (Fig. 8).
Effect of various nitrogen sources
Supplementation of various nitrogen sources i.e., yeast extract, malt extract and peptone to medium was also studied. The best results were obtained with yeast extract for enzyme production (125.60±2.17 U/mL) while lowest synthesis was observed with malt extract (Fig. 9).
Evaluation of optimal concentration of media components
In the following investigation, CCD of RSM was used for the prediction of medium composition and to maximize the tannase production from R. ornithinolytica in SSF. Tannase production was observed from 27.584 U/ml to 157.041 U/mL in 17 fermentation runs of CCD method (Table III). The model was significant having F and P values of 4.605148 and 0.028247 respectively (Table IV). There was slight difference between observed and predicted values (Fig. 10) which also showed model accuracy. Maximum tannase production (157.0414 U/mL) was attained at run number 3, where components were: 3.75% tannic acid, 0.75% K2HPO4 and 1.25% yeast extract. The response of the design was calculated using second order polynomial regression equation.
Y (trannase production) = 1208.718 – 790.189A – 132.044B – 946.171C + 94.638A2 + 38.280 B2 + 5.084 C2 – 36.955A*B + 537.251A*C + 186.439B*C
After data analysis, contour plots (Fig. 11) were constructed which indicated that each parameter had significant effect on tannase production. Figure 12 represented desirability chart for tannase production which revealed that the results were verified by repeated experiments.
Characterization of tannase activity
Effect of pH
Experiments were performed to observe the pH effect on tannase activity by applying pH ranges 4.0 to 11.0. Results (Fig. 13) indicated a reasonable enzyme activity in this pH range with significantly higher activity (168.41±0.36 U/mL) at pH 5. Furthermore, the activity was observed to be decreased with the increase of pH after optimum point.
Effect of incubation temperature
Incubation temperature ranging from 20 to 90°C was applied to determine its effect on tannase activity. Tannase from R. ornithinolytica was significantly higher at 30°C, making it optimum temperature for tannase activity of 190.14±1.13 U/mL (Fig. 13).
Table IV. ANOVA values for regression model obtained from CCD applied for medium component optimization for tannase production from R. ornithinolytica in SSF.
|
Effect |
SS |
DF |
MS |
F-value |
P-value |
|
Model |
13825.58 |
9 |
1536.176 |
4.605148 |
0.028247 |
|
A |
3949.293 |
1 |
3949.293 |
11.83919 |
0.010825 |
|
A2 |
688.338 |
1 |
688.338 |
2.06350 |
0.194011 |
|
B |
138.374 |
1 |
138.374 |
0.41482 |
0.540056 |
|
B2 |
78.342 |
1 |
78.342 |
0.23485 |
0.642745 |
|
C |
6728.790 |
1 |
6728.790 |
20.17156 |
0.002828 |
|
C2 |
1.986 |
1 |
1.986 |
0.00595 |
0.940654 |
|
AB |
42.678 |
1 |
42.678 |
0.12794 |
0.731119 |
|
AC |
9019.952 |
1 |
9019.952 |
27.04001 |
0.001253 |
|
BC |
1086.239 |
1 |
1086.239 |
3.25633 |
0.114125 |
|
Error |
2335.046 |
7 |
333.578 |
A, tannic acid; B, K2PHO4; C, yeast extract; SS, sum of squares, MS, mean square; DF, degrees of freedom.
Effect of incubation time
The activity of activity was investigated at several incubation times (15, 30, 45…, 175 minutes) to determine the optimum incubation time. The highest activity (194.07±0.11 U/mL) was detected at incubation of 60 minutes (Fig. 13). After optimum time, the tannase activity kept on decreasing.
Effect of substrate concentration
Tannase activity was studied at different substrate concentrations (0.25, 0.3, 0.35…, 0.6%). The maximum tannase activity (203.52±0.62 U/mL) resulted at 0.45% of substrate (Fig. 13).
DISCUSSION
In present study, tannase producing potential of already isolated bacterial strains from fish gut content were screened on the medium supplemented with 0.5% tannic aicd. Previously, few investigations have been reported to evaluate the tannase producing capability of fish gut microbes. Mandal and Ghosh (2013a) described the isolation of tannase-producing microbiota from gut of freshwater fishes using tannic acid incorporated selective medium. Similarly, Talukdar et al. (2016) reported the isolation of many tannase producer bacteria from gastrointestinal tract of seven different fishes. For screening of tannase producing microorganisms, Brahmbhatt et al. (2014) also used nutrient-agar medium incorporated with tannic acid (0.5%) similar to our investigation. It is now well-known that tannins present in forages cause the retardation of productivity and growth in grazing animals (Goel et al., 2005). In herbivorous and omnivorous fishes, adverse effects have been observed by tannin-rich feed (Becker and Makkar, 1999). The tannin-degrading microbiota in gut might be as the outcome of coevolution of tannin compounds and omnivorous/herbivorous fishes (Mandal and Ghosh, 2013a). On the basis of large zone and enzyme assay, Raoultella ornithinolytica was selected. R. ornithinolytica showed enzyme production up to 3.31 U/mL on growth medium (CaCl2 0.1%, yeast extract 0.275%, tannic acid 0.5%) as used by Javed (2016) previously in submerged fermentation. Sivashanmugam and Jayaraman (2011) reported 3.9 U/ml tannase production with 1% tannic acid, 0.05% KCl, 0.05% MgSO4, 0.1 % K2HPO4, and 3% NaNO3. For Bacillus licheniformis, tannase production (0.356 U/ml) was reported with medium comprising of tannic acid 1.0%, NH4Cl 0.35%, KH2PO40.45%, and MgSO4 0.05% (Mohapatra et al., 2009). These reports indicate that different bacterial strains have different tannase production potential that may be affected by the selection of growth medium components and concentrations.
Moisture content has significant effect during SSF and its value depends on microorganism as well as substrate used (Kalogeris et al., 2003). Certain water quantity is required for synthesis of new cells. The substrate bulging is also caused by moisture that facilitate microbial action (Pandey et al., 2000; Sabu et al., 2006). In our study, tannase production increased with the increase of initial moisture content up to 60%. Afterward, the decline in tannase synthesis was observed with the rise of moisture value. Mandal and Ghosh (2013b) also reported the similar trend with optimal tannase synthesis at 60% moisture in groundnut oil cake substrate. In Sabu et al. (2006) investigation, the optimal tannase synthesis from Lactobacillus sp. ASR-S1 was achieved with 50% moisture in coffee husk while production tend to decline after optimal level. Lesser enzyme at higher value of moisture might be because of lower oxygen supply leading to lower biomass and enzyme synthesis (Manjit et al., 2008). At very lower and higher moisture content, the organic matter degradation becomes lower, that consequently affects the enzyme production (Pandey et al., 2001). During current study, tap water was found to be optimal extraction medium among distilled water, tap water, 1% NaCl, acetate buffer of pH 4.0 and 5.0, phosphate buffer of pH 6.0 and 7.0, tris-HCl buffer of pH 8 and 9 and glycine NaOH buffer of pH 10 and 11. Tannase is mostly extracellular and can be extracted using water as well as buffer (Aguilar et al., 2007). Chatterjee et al. (1996) used water for the extraction of tannase. While Sabu et al. (2006) reported 0.05 M citrate buffer (pH 5) as extraction medium for tannase production.
Temperature is an important parameter during enzyme synthesis that may cause the protein denaturation, inhibition or promotion of specific metabolites, inhibition of enzyme and even the cell death (Sabu et al., 2006). In this study, the production of tannase increased with rising temperature and optimal production value was recorded at 45°C. Few reports demonstrated the tannase synthesis at high temperature. Optimum temperature for tannase production in SSF, in general, falls in 25–35°C range (Aharwar and Parihar, 2018). Aftab et al. (2016) also reported the increase in tannase synthesis with increasing incubation temperature up to 41°C where maximum tannase production from Bacillus subtilis was observed. Such variations in optimal temperature of incubation could be due to differences in microorganisms’ nature and their environment. Experiments on optimizing pH for tannase production in SSF revealed the tannase synthesis increased with increasing pH till pH 7.0. With further pH increase, decline in enzyme production was recorded. Talukdar et al. (2016) also observed the neutral pH as optimum for tannase production from strains isolated from fish gut rather than alkaline or acidic pH. Whereas, Aharwar and Parihar (2018) documented that in SSF, tannase production, in general, is optimal in acidic pH. The neutral pH for optimal tannase synthesis might be due to the adaptation of bacterial strains in the neutral/alkaline environment of gastrointestinal tract of fish (Talukdar et al., 2016). Khan and Ghosh (2013) also reported the neutral pH as the most favorable for enzyme production form Bacillus subtilis isolated from fish gut. With the increase of inoculum size R. ornithinolytica from 1% to 3%, the enzyme production was recorded to be increased and highest tannase was produced when 3% inoculum was added. In SSF, the inoculum size plays a vital role for metabolite synthesis. Lower size of inoculum (number of cells) would not be enough for bacterial biomass and synthesis of enzyme, so large size inoculum is required for proper enzyme yield (Kashyap et al., 2002; Sabu et al., 2006). However, Banerjee et al. (2007) showed the optimal growth with 2% inoculum of Aureobasidium pullulans DBS66 while Beniwal et al. (2010) observed the maximum tannase using 1% Enterobacter cloacae MTCC 9125 inoculum in SSF.
The R. ornithinolytica showed the maximum tannase production when 24 h of incubation was given and with further increase of time caused the decreased tannase. Our results show good agreement with Mondal et al. (2001) who also detected the highest tannase level produced by Bacillus cereus KBR9 at 24 h of incubation after which its level decreased. With the increase of incubation period, production of enzyme deceased that could be the result of denaturation and inhibition of enzyme with the time (Gautam et al., 2002; Paranthaman et al., 2010). Exhaustion of substrate and elevation of byproducts with the time could also decline the enzyme production (Souza et al., 2018). However, Jana et al. (2013) reported the optimal tannase synthesis after 72 h incubation. During present investigation, agitation (150 rpm) gave better results rather than static condition. Similar outcomes were stated by Murad et al. (2014) who suggested agitation an important parameter for dissolving oxygen in the medium. Kumar et al. (2015b) reported the highest tannase production with agitation at 103.34 rpm. The Enterobacter cloacae strain 41 was reported to give maximum tannase yield with agitation at 100 rpm (Govindarajan et al., 2019). Agitation affects significantly on the fermentation as it promotes proper blending of medium and mixing of oxygen (Darah et al., 2011). However, Serratia marcesans showed higher tannase value at aerobic static condition (Sheela et al., 2016). The impact of size of substrate particles on tannase production was evaluated and particles with greater diameter yielded the best results. Comparable results were described by Yee et al. (2011) for tananse synthesis in SSF. More enzyme is produced by using larger substrate particles which facilitate more aeration and higher respiration leading to better reactions (John et al., 2006). Smaller particles may agglomerate that reduce the surface area for microbial action and lower the enzyme production (Krishna, 2005). However, Madeira Jr et al. (2015) reported the higher tannase production with small sized particles of substrate.
Like other enzymes, metal ions are also required for enhanced microbial growth and tannase biosynthesis as well as for proper catalytic activity (Jana et al., 2014). Different metal salts may affect tannase production differently. Among different salts, addition of KH2PO4 enhanced the enzyme production. Wu et al. (2018) also reported the enhanced microbial biomass and tannase production with the addition of 0.1% K2HPO4 in the medium. During tannase production, KH2PO4 might be act as phosphate source as mostly metabolisms require phosphorylation of their respective proteins that is responsible for enzyme activation as well as inactivation. Moreover, KH2PO4 is responsible for maintaining buffer situation in medium (Jana et al., 2014). Tannase from microbial source is an inducible enzyme, therefore require tannin or tannic acid as an inducer for its synthesis (Mansor et al., 2019). When different concentrations of tannic acid (1.5 to 4.0%) were applied, the maxium tannase was produced by R. ornithinolytica using 3.5% tannic acid while with further rise of its concentration the decline in enzyme synthesis was recorded. In agreement to our result, Seth and Chand (2000) also found 3.5% tannic acid as optimum for tannase synthesis. The decline of tannase synthesis in higher concentration could be due to toxicity of substrate or as a result of by products (gallic acid and glucose) accumulation on membrane or inhibition of substrate (Seth and Chand, 2000; Beniwal et al., 2010). Another reason could be the binding of gallic acid to active site by mimicking the substrate and thus blocking of enzyme action is caused that lower the enzyme production (Kar et al., 1999; Kumar et al., 1999). For tannase production, nitrogen source is considered to be a vital factor which has effect over microbial biomass and enzyme synthesis (Patel et al., 2005). Most of microorganisms use nitrogen source for the synthesis of amino acids, protein, components of cell wall and nucleic acids (Jana et al., 2013). In our study, the maximum enzyme was produced when organic nitrogen source was given as yeast extract. Comparable results were observed by Murad et al. (2014) and Reddy and Kumar (2012) who also reported best results with yeast extract among all organic nitrogen source supplied. While in contrast, Battestin and Macedo (2007) and Govindarajan et al. (2019) described the negative impact of yeast extract over tannase production.
Concentration of medium components i.e., tannic acid, K2HPO4 and yeast extract were optimized by CCD of RSM to maximize tannase production from R. ornithinolytica using corn residues. With 3.75% tannic acid, 0.75% K2HPO4 and 1.25% yeast extract highest enzyme production (157.0414 U/mL) was obtained.Lima et al. (2014) obtained optimum tannase production with 3.5% tannic acid concentration during RSM using Barbados cherry as substrate. While Mohan et al. (2014) used 3.22 % tannic acid during RSM for optimal tannase synthesis. For tannase synthesis, 1% tannic acid was found to most significant using Streptomyces sp. AT 13 in RSM (Tripathi and Lakshmi, 2018). However, highest tannase was produced using 6% tannic acid in Madeira et al. (2011) investigation. Wu et al. (2018) determined optimum tannase using 2.25% yeast extract during RSM.
Effect of different parameters i.e., pH, temperature, time of incubation and substrate concentrations over tannase activity was determined. The pH largely affects the enzyme reaction as it influences of acidic and basic amino acid’s ionization state (Jana et al., 2014). Effect of different pH on tannase activity depict pH 5.0 as optimum pH while activity tend to decrease at higher pH with the lowest activity at pH 11.0. In acidic pH range (4.0-6.0), tannase exhibited the higher activity. Our results were in agreement with tannase activity of Lactobacillus plantarum with optima at 5.0 pH (Rodríguez et al., 2008). Tannase is an acidic protein having higher activity generally in acidic range (Jana et al., 2014). Optimal pH for tannase activity was reported from 5.0 to 6.0 by many authors (Gayen and Ghosh, 2013; Bagga et al., 2015; Kumar et al., 2015; Farag et al., 2018). However, tannase activity in alkaline range has also been reported (Iwamoto et al., 2008).
Tannase from R. ornithinolytica was significantly higher at 30°C, tend to reduce after this temperature. Equivalent results were found by Lopes et al. (2018) from Saccharomyces cerevisiae tannase with decreasing activity after temperature optima. Rodríguez et al. (2008) and Nadaf and Ghosh (2011) also reported tannase activity for Lactobacillus plantarum and Rhodococcus NCIM 2891 respectively with optimum temperature of 30°C. Mostly, tannase activity has temperature optima in mesophilic range (Jana et al., 2014). With the increase of temperature of kinetic energy of substrate and enzymes elevates that facilitates the enzyme reaction. After optimum level, the chemical potential energy becomes so high that weak bonds involved in three-dimensional structure break down and thus denaturation and inactivation of substrate or enzyme molecules are caused (Mukherjee and Banerjee, 2006). However, Enterobacter sp. and Klebsiella pneumoniae tananses activity was maximum at 40°C and 50°C respectively (Sharma and John, 2011; Kumar et al., 2015a).
The impact of incubation period over tannase activity depicted the maxium value after 60 minutes incubation and by further increasing the time period, the activity decreased. The decrease in activity may be because of enzyme denaturation with time (Gautam et al., 2002). The activity of tannase was improved by raising the concentration of tannic acid up to 0.45% tannic acid, where highest tannase activity was recorded. Beyond the optimum level, tannase activity tend to decline with the increase of tannic acid substrate.
CONCLUSION
Tannase or specifically tannin acyl hydrolase EC 3.1.1.20 is an economically valuable enzyme with applications in food, beverages, pharmaceutical, cosmetics, leather and chemical production. The use of an expensive substrate is a big challenge for industrial viewpoint. Our results support that the bacterial strain R. ornithinolytica isolated from fish gut content had a great potential to utilize low-cost agricultural residues of corn (Zea mays) as substrate for tannase production. Optimization of physical parameters and medium components is essential to maximize the enzyme production and activity at large-scale level.
Statement of conflict of interest
The authors have declared no conflict of interest.
REFERENCES
Aftab, M.N., Mukhtar, H. and Haq, I., 2016. Production and characterization of tannase from a newly isolated Bacillus subtilis. Pak. J. Bot., 48: 1263-1271.
Aguilar, C.N. and Gutiérrez-Sánchez, G., 2001. Sources, properties, applications and potential uses of tannin acyl hydrolase. Fd. Sci. Technol. Int., 7: 373-382. https://doi.org/10.1106/69M3-B30K-CF7Q-RJ5G
Aguilar, C.N., Augur, C., Favela-Torres, E. and Viniegra-González, G., 2001. Induction and repression patterns of fungal tannase in solid-state and submerged cultures. Process Biochem., 36: 565-570. https://doi.org/10.1016/S0032-9592(00)00251-X
Aguilar, C.N., Favela-Torres, E., Vinegra-González, G. and Augur, C., 2002. Culture conditions dictate protease and tannase production in submerged and solid-state cultures of Aspergillus niger Aa-20. Appl. Biochem. Biotechnol., 102: 407-414. https://doi.org/10.1385/ABAB:102-103:1-6:407
Aguilar, C.N., Rodríguez, R., Gutiérrez-Sánchez, G., Augur, C., Favela-Torres, E., Prado-Barragan, L.A., Ramírez-Coronel, A. and Contreras-Esquivel, J.C., 2007. Microbial tannases: Advances and perspectives. Appl. Microbiol. Biotechnol., 76: 47-59. https://doi.org/10.1007/s00253-007-1000-2
Aharwar, A. and Parihar, D.K., 2018. Tannases: Production, properties, applications. Biocatal. Agric. Biotechnol., 15: 322-334. https://doi.org/10.1016/j.bcab.2018.07.005
Aissam, H., Errachidi, F., Penninckx, M.J., Merzouki, M. and Benlemlih, M., 2005. Production of tannase by Aspergillus niger HA37 growing on tannic acid and olive mill waste waters. World J. Microbiol. Biotechnol., 21: 609-614. https://doi.org/10.1007/s11274-004-3554-9
Ayed, L. and Hamdi, M., 2002. Culture conditions of tannase production by Lactobacillus plantarum. Biotechnol. Lett., 24: 1763-1765. https://doi.org/10.1023/A:1020696801584
Bagga, J., Pramanik, S.K. and Pandey, V., 2015. Production and purification of tannase from Aspergillus aculeatus using plant derived raw tannin. Int. J. Sci. Eng. Technol., 4: 50-55. https://doi.org/10.17950/ijset/v4s2/205
Banerjee, D., Mondal, K. and Pati, B., 2007. Tannase production by Aspergillus aculeatus DBF9 through solid-state fermentation. Acta Microbiol. Imm. H., 54: 159-166. https://doi.org/10.1556/AMicr.54.2007.2.6
Barrios-González, J., 2012. Solid-state fermentation: physiology of solid medium, its molecular basis and applications. Process Biochem., 47: 175-185. https://doi.org/10.1016/j.procbio.2011.11.016
Battestin, V. and Macedo, G.A., 2007. Tannase production by Paecilomyces variotii. Bioresour. Technol., 98: 1832-1837. https://doi.org/10.1016/j.biortech.2006.06.031
Becker, K. and Makkar, H.P.S., 1999. Effects of dietary tannic acid and quebracho tannin on growth performance and metabolic rates of common carp (Cyprinus carpio L.). Aquaculture, 175: 327-335. https://doi.org/10.1016/S0044-8486(99)00106-4
Beena, P.S., Basheer, S.M., Bhat, S.G., Bahkali, A.H. and Chandrasekaran, M., 2011. Propyl gallate synthesis using acidophilic tannase and simultaneous production of tannase and gallic acid by marine Aspergillus awamori BTMFW032. Appl. Biochem. Biotechnol., 164: 612-628. https://doi.org/10.1007/s12010-011-9162-x
Beniwal, V., Chhokar, V., Singh, N. and Sharma, J., 2010. Optimization of process parameters for the production of tannase and gallic acid by Enterobacter cloacae MTCC 9125. J. Am. Sci., 6: 389-397.
Böer, E., Bode, R., Mock, H.P., Piontek, M. and Kunze, G., 2009. Atan1 pan extracellular tannase from the dimorphic yeast Arxula adeninivorans: Molecular cloning of the ATAN1 gene and characterization of the recombinant enzyme. Yeast, 26: 323-337. https://doi.org/10.1002/yea.1669
Böer, E., Breuer, F.S., Weniger, M., Denter, S., Piontek, M. and Kunze, G., 2011. Large-scale production of tannase using the yeast Arxula adeninivorans. Appl. Microbiol. Biotechnol., 92: 05. https://doi.org/10.1007/s00253-011-3320-5
Brahmbhatt, D., Modi, H.A. and Jain, N.K., 2014. Preliminary isolation and screening of tannase producing bacteria and fungi. Int. J. Curr. Microbiol. appl. Sci., 3: 193-203.
Cannel, E., 1980. Solid-state fermentation sys-tems. Process Biochem., 15: 2-7.
Chandrasekaran, M., 1991. Combined effect of environmental factors on spoilage bacteria. Fish Technol., 28: 146-153.
Chatterjee, R., Dutta, A., Banerjee, R. and Bhattacharyya, B.C., 1996. Production of tannase by solid-state fermentation. Bioproc. Eng., 14: 159-162. https://doi.org/10.1007/BF00369434
Chavez, C., 1988. The use of seed of the leguminous plant Sesbania grandiflora as a partial replacement for fish meal in diets for tilapia (Oreochromis mossambicus). Aquaculture, 71: 51-60. https://doi.org/10.1016/0044-8486(88)90272-4
Chávez-González, M.L., Guyot, S., Rodríguez-Herrera, R., Prado-Barragán, A. and Aguilar, C.N., 2014. Production profiles of phenolics from fungal tannic acid biodegradation in submerged and solid-state fermentation. Process Biochem., 49: 541-546. https://doi.org/10.1016/j.procbio.2014.01.031
Darah, I., Sumathi, G., Jain, K. and Lim, S.H., 2011. Influence of agitation speed on tannase production and morphology of Aspergillus niger FETL FT3 in submerged fermentation. Appl. Biochem. Biotechnol., 165: 1682-1690. https://doi.org/10.1007/s12010-011-9387-8
de Brito, A.R., Reis, N.S., Silva, T.P., Bonomo, R.C.F., Uetanabaro, A.P.T., de Assis, S.A., da Silva, E.G.P., Aguiar-Oliveira, E., Oliveira, J.R. and Franco, M., 2017. Comparison between the univariate and multivariate analysis on the partial characterization of the endoglucanase produced in the solid state fermentation by Aspergillus oryzae ATCC 10124. Prep. Biochem. Biotechnol., 47: 977-985. https://doi.org/10.1080/10826068.2017.1365247
Esakkiraj, P., Immanuel, G., Sowmya, S.M., Iyapparaj, P. and Palavesam, A., 2009. Evaluation of protease-producing ability of fish gut isolate Bacillus cereus for aqua feed. Fd. Bioproc. Tech., 2: 383. https://doi.org/10.1007/s11947-007-0046-6
Farag, A.M., Hassan, S.W., Asmaa, M. and Ghanem, K.M., 2018. Purification, characterization and application of tannase enzyme isolated from marine Aspergillus nomius GWA5. J. Pure appl. Microbiol., 12: 1939-1950. https://doi.org/10.22207/JPAM.12.4.30
Ferraz, J.L.A.A., Souza, L.O., Fernandes, A.G.A., Oliveira, M.L.F., de Oliveira, J.R. and Franco, M., 2020. Optimization of the solid-state fermentation conditions and characterization of xylanase produced by Penicilliumroqueforti ATCC 10110 using yellow mombin residue (Spondias mombin L.). Chem. Eng. Commun., 207: 31-42. https://doi.org/10.1080/00986445.2019.1572000
Gautam, P., Sabu, A., Pandey, A., Szakacs, G. and Soccol, C.R., 2002. Microbial production of extra-cellular phytase using polystyrene as inert solid support Bioresour. Technol., 83: 229-233. https://doi.org/10.1016/S0960-8524(01)00215-2
Gayen, S. and Ghosh, U., 2013. Purification and characterization of tannin acyl hydrolase produced by mixed solid state fermentation of wheat bran and marigold flower by Penicillium notatum NCIM 923. Biomed. Res. Int., 2013. https://doi.org/10.1155/2013/596380
Goel, G., Puniya, A.K., Aguilar, C.N. and Singh, K., 2005. Interaction of gut microflora with tannins in feeds. Naturwissenschaften, 92: 497-503. https://doi.org/10.1007/s00114-005-0040-7
Govindarajan, R.K., Krishnamurthy, M., Neelamegam, R., Shyu, D.J., Muthukalingan, K. and Nagarajan, K., 2019. Purification, structural characterization and biotechnological potential of tannase enzyme produced by Enterobacter cloacae strain 41. Process Biochem., 77: 37-47. https://doi.org/10.1016/j.procbio.2018.10.013
Govindarajan, R.K., Revathi, S., Rameshkumar, N., Krishnan, M. and Kayalvizhi, N., 2016. Microbial tannase: Current perspectives and biotechnological advances. Biocatal. Agric. Biotechnol., 6: 168-175. https://doi.org/10.1016/j.bcab.2016.03.011
Iwamoto, K., Tsuruta, H., Nishitaini, Y. and Osawa, R., 2008. Identification and cloning of a gene encoding tannase (tannin acylhydrolase) from Lactobacillus plantarum ATCC 14917T. Syst. appl. Microbiol., 31: 269-277. https://doi.org/10.1016/j.syapm.2008.05.004
Jana, A., Halder, S.K., Banerjee, A., Paul, T., Pati, B.R., Mondal, K.C. and Mohapatra, P.K.D., 2014. Biosynthesis, structural architecture and biotechnological potential of bacterial tannase: A molecular advancement. Bioresour. Technol., 157: 327-340. https://doi.org/10.1016/j.biortech.2014.02.017
Jana, A., Maity, C., Halder, S.K., Das, A., Pati, B.R., Mondal, K.C. and Mohapatra, P.K.D., 2013. Structural characterization of thermostable, solvent tolerant, cytosafe tannase from Bacillus subtilis PAB2. Biochem. Eng. J., 77: 161-170. https://doi.org/10.1016/j.bej.2013.06.002
Javed, I., 2016. Evaluation of tannase producing potential of bacteria isolate from fish gut. BS thesis, Department of Zoology, University of the Punjab, Lahore, Pakistan
John, R.P., Nampoothiri, K.M. and Pandey, A., 2006. Solid-state fermentation for L-lactic acid production from agro wastes using Lactobacillus delbrueckii. Process Biochem., 41: 759-763. https://doi.org/10.1016/j.procbio.2005.09.013
Kalogeris, E., Iniotaki, F., Topakas, E., Christakopoulos, P., Kekos, D. and Macris, B.J., 2003. Performance of an intermittent agitation rotating drum type bioreactor for solid-state fermentation of wheat straw. Bioresour. Technol., 86: 207-213. https://doi.org/10.1016/S0960-8524(02)00175-X
Kar, B., Banerjee, R. and Bhattacharyya, B.C., 1999. Microbial production of gallic acid by modified solid state fermentation. J. ind. Microbiol. Biotechnol., 23: 73-177. https://doi.org/10.1038/sj.jim.2900713
Kashyap, P., Sabu, A., Pandey, A., Szakacs, G. and Soccol, C.R., 2002. Extra-cellular L-glutaminase production by Zygosaccharomyces rouxii under solid-state fermentation. Process Biochem., 38: 307-312. https://doi.org/10.1016/S0032-9592(02)00060-2
Khan, A., and Ghosh, K., 2013. Evaluation of phytase production by fish gut bacterium, Bacillus subtilis, for processing of Ipomea aquatica leaves as probable aquafeed ingredient. J. Aquat. Fd. Prod. Technol., 22: 508-519. https://doi.org/10.1080/10498850.2012.669032
Khuri, A.I. and Mukhopadhyay, S., 2010. Response surface methodology. Wiley Interdiscipl. Rev.Comput. Stat., 2: 128-149. https://doi.org/10.1002/wics.73
Krishna, C., 2005. Solid-state fermentation systems. an overview. Crit. Rev. Biotechnol., 25: 1-30. https://doi.org/10.1080/07388550590925383
Kumar, M., Beniwal, V. and Salar, R.K., 2015. Purification and characterization of a thermophilic tannase from Klebsiella pneumoniae KP715242. Biocatal Agric. Biotechnol., 4: 745-751. https://doi.org/10.1016/j.bcab.2015.10.011
Kumar, M., Rana, S., Beniwal, V. and Salar, R.K., 2015. Optimization of tannase production by a novel Klebsiella pneumoniae KP715242 using central composite design. Biotechnol. Rep., 7: 128-134. https://doi.org/10.1016/j.btre.2015.06.002
Kumar, R., Sharma, J. and Singh, R., 2007. Production of tannase from Aspergillus ruber under solid-state fermentation using jamun (Syzygium cumini) leaves. Microbiol. Res., 162: 384-390. https://doi.org/10.1016/j.micres.2006.06.012
Kumar, R.A., Gunasekaran, P. and Lakshmanan, M., 1999. Biodegradation of tannic acid by Citrobacter freundii isolated from a tannery effluent. J. Basic Microbiol., 39: 161-168. https://doi.org/10.1002/(SICI)1521-4028(199906)39:3<161::AID-JOBM161>3.0.CO;2-U
Lessa, O.A., Reis, N.D.S., Leite, S.G.F., Gutarra, M.L.E., Souza, A.O., Gualberto, S.A., de Oliveira, J.R., Aguiar-Oliveira, E. and Franco, M., 2018. Effect of the solid state fermentation of cocoa shell on the secondary metabolites, antioxidant activity, and fatty acids. Fd. Sci. Biotechnol., 27: 107–113. https://doi.org/10.1007/s10068-017-0196-x
Lima, J.S.D., Cruz, R., Fonseca, J.C., Medeiros, E.V.D., Maciel, M.D.H.C., Moreira, K.A. and Motta, C.M.D.S., 2014. Production, characterization of tannase from Penicillium montanense URM 6286 under SSF using agroindustrial wastes, and application in the clarification of grape juice (Vitis vinifera L.). Sci. World J., 2014. https://doi.org/10.1155/2014/182025
Liu, T.P., Porto, T.S., Moreira, K.A., Takaki, G.M., Brandão-Costa, R., Herculano, P.N. and Porto, A.L., 2016. Tannase production by Aspergillus spp. UCP1284 using cashew bagasse under solid state fermentation. Afr. J. Microbiol. Res., 10: 565-571. https://doi.org/10.5897/AJMR2016.7924
Lokeswari, N. and Kumar, L.N., 2013. Tannase production from cashew husk by solid state fermentation. Int. J. Adv. Biol. Res., 3: 295-299.
Lopes, L.M.D.M., Costa Batista, L.H., Gouveia, M.J., Leite, T.C.C., de Mello, M.R.F., de Assis, S.A. and de Sena, A.R., 2018. Kinetic and thermodynamic parameters, and partial characterization of the crude extract of tannase produced by Saccharomyces cerevisiae CCMB 520. Nat. Prod. Res., 32: 1068-1075. https://doi.org/10.1080/14786419.2017.1380010
Madeira Jr, J.V., Ferreira, L.R., Macedo, J.A. and Macedo, G.A., 2015. Efficient tannase production using Brazilian citrus residues and potential application for orange juice valorization. Biocatal. Agric. Biotechnol., 4: 91-97. https://doi.org/10.1016/j.bcab.2014.11.005
Madeira Jr, J.V., Macedo, J.A. and Macedo, G.A., 2011. Detoxification of castor bean residues and the simultaneous production of tannase and phytase by solid-state fermentation using Paecilomyces variotii. Bioresour. Technol., 102: 7343-7348. https://doi.org/10.1016/j.biortech.2011.04.099
Mandal, S. and Ghosh, K., 2013a. Isolation of tannase-producing microbiota from the gastrointestinal tracts of some freshwater fish. J. appl. Ichthyol., 29: 145-153. https://doi.org/10.1111/j.1439-0426.2012.02054.x
Mandal, S. and Ghosh, K., 2013b. Optimization of tannase production and improvement of nutritional quality of two potential low-priced plant feedstuffs under solid state fermentation by Pichia kudriavzevii isolated from fish gut. Fd. Biotechnol., 27: 86-103. https://doi.org/10.1080/08905436.2012.755929
Manjit, K.S., Yadav, A., Aggarwal, N.K., Kumar, K. and Kumar, A., 2008. Tannase production by Aspergillus fumigatus MA under solid-state fermentation. World J. Microbiol. Biotechnol., 24: 3023-3030. https://doi.org/10.1007/s11274-008-9847-7
Mansor, A., Ramli, M.S., Rashid, N.A., Samat, N., Lani, M.N., Sharifudin, S.A. and Raseetha, S., 2019. Evaluation of selected agri-industrial residues as potential substrates for enhanced tannase production via solid-state fermentation. Biocatal. Agric. Biotechnol., 20: 101216. https://doi.org/10.1016/j.bcab.2019.101216
Miller, G.L., 1959. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem., 31: 426-428. https://doi.org/10.1021/ac60147a030
Mohan, S.K., Viruthagiri, T. and Arunkumar, C., 2014. Statistical optimization of process parameters for the production of tannase by Aspergillus flavus under submerged fermentation. 3 Biotech, 4: 159-166. https://doi.org/10.1007/s13205-013-0139-z
Mohapatra, P.D., Maity, C., Rao, R.S., Pati, B.R. and Mondal, K.C., 2009. Tannase production by Bacillus licheniformis KBR6: Optimization of submerged culture conditions by Taguchi DOE methodology. Fd. Res. Int., 42: 430-435. https://doi.org/10.1016/j.foodres.2009.02.013
Mondal, K.C., Banerjee, D., Banerjee, R. and Pati, B.R., 2001. Production and characterization of tannase from Bacillus cereus KBR9. J. Gen. appl. Microbiol., 47: 263-267. https://doi.org/10.2323/jgam.47.263
Mukherjee, G. and Banerjee, R., 2006. Effects of temperature, pH and additives on the activity of tannase produced by a co-culture of Rhizopus oryzae and Aspergillus foetidus. World J. Microbiol. Biotechnol., 22: 207-212. https://doi.org/10.1007/s11274-005-9022-3
Murad, H.A., El Tawab, A.A., Kholif, A.M., El-Nor, S.A., Matloup, O.H., Khorshed, M.M. and El-Sayed, H.M., 2014. Production of tannase by Aspergillus niger from palm kernel. Biotechnology, 13: 68-73. https://doi.org/10.3923/biotech.2014.68.73
Muslim, S.N., Mahammed, A.N., Musafer, H.K., Al-Kadmy, I.M., Shafiq, S.A. and Muslim, S.N., 2015. Detection of the optimal conditions for tannase productivity and activity by Erwinia Carotovora. J. med. Bioeng., 4: 198-205. https://doi.org/10.12720/jomb.4.3.198-205
Nadaf, N.H. and Ghosh, J.S., 2011. Production, purification and characterization of tannase from Rhodococcus NCIM, 2891. Curr. Res. J. biol. Sci., 3: 246-253.
Osawa, R. and Walsh, T.P., 1993. Visual reading method for detection of bacterial tannase. Appl. environ. Microbiol., 59: 1251-1252. https://doi.org/10.1128/AEM.59.4.1251-1252.1993
Pandey, A., 1994. Solid state fermentation: An overview. In: Solid state fermentation (ed. A. Pandey). Wiley Eastern Limited, New Delhi, pp. 3–10.
Pandey, A., Soccol, C.R., Nigam, P. and Soccol, V.T., 2000. Biotechnological potential of agro-industrial residues. I: sugarcane bagasse. Bioresour. Technol., 74: 69-80. https://doi.org/10.1016/S0960-8524(99)00142-X
Pandey, A., Soccol, C.R., Rodriguez-Leon, J.A. and Nigam, P.S.N., 2001. Solid state fermentation in biotechnology: Fundamentals and applications. Reference book.
Paranthaman, R., Vidyalakshmi, R., Murugesh, S. and Singaravadivel, K., 2010. Manipulation of fermentation conditions on production of tannase from agricultural by-products with Aspergillus oryzae. Afr. J. Microbiol. Res., 4: 1440-1445.
Patel, R., Dodia, M. and Singh, S.P., 2005. Extracellular alkaline protease from a newly isolated haloalkaliphilic Bacillus sp.: Production and optimization. Process Biochem., 40: 3569-3575. https://doi.org/10.1016/j.procbio.2005.03.049
Raghuwanshi, S., Dutt, K., Gupta, P., Misra, S. and Saxena, R.K., 2011. Bacillus sphaericus: The highest bacterial tannase producer with potential for gallic acid synthesis. J. Biosci. Bioeng., 111: 635-640. https://doi.org/10.1016/j.jbiosc.2011.02.008
Reddy, M.N. and Kumar, C.G., 2012. Production oftannase by isolated Aspergillus terreus under solid state fermentation. Int. J. Pharma. Res. Dev., 3: 41-49.
Rodríguez, H., de las Rivas, B., Gómez-Cordovés, C. and Muñoz, R., 2008. Characterization of tannase activity in cell-free extracts of Lactobacillus plantarum CECT 748T. Int. J. Fd. Microbiol., 121: 92-98. https://doi.org/10.1016/j.ijfoodmicro.2007.11.002
Sabu, A., Augur, C., Swati, C. and Pandey, A., 2006. Tannase production by Lactobacillus sp. ASR-S1 under solid-state fermentation. Process Biochem., 41: 575-580. https://doi.org/10.1016/j.procbio.2005.05.011
Sabu, A., Pandey, A., Daud, M.J. and Szakacs, G., 2005. Tamarind seed powder and palm kernel cake: two novel agro residues for the production of tannase under solid state fermentation by Aspergillus niger ATCC 16620. Bioresour. Technol., 96: 1223-1228. https://doi.org/10.1016/j.biortech.2004.11.002
Santos, T.C.D., Filho, G.A., De Brito, A.R., Pires, A.J.V., Bonomo, R.C.F., and Franco, M., 2016. Production and characterization of cellulolytic enzymes by Aspergillus niger and Rhizopus sp. by solid state fermentation of prickly pear. Rev. Caatinga, 29: 222–233. https://doi.org/10.1590/1983-21252016v29n126rc
Selwal, M.K., Yadav, A., Selwal, K.K., Aggarwal, N.K., Gupta, R. and Gautam, S.K., 2010. Optimization of cultural conditions for tannase production by Pseudomonas aeruginosa IIIB 8914 under submerged fermentation. World J. Microbiol. Biotechnol., 26: 599-605. https://doi.org/10.1007/s11274-009-0209-x
Seth, M. and Chand, S., 2000. Biosynthesis of tannase and hydrolysis of tannins to gallic acid by Aspergillus awamori-optimisation of process parameters. Process Biochem., 36: 39-44. https://doi.org/10.1016/S0032-9592(00)00179-5
Sharma, K.P. and John, P.J., 2011. Purification and characterization of tannase and tannase gene from Enterobacter sp. Process Biochem., 46: 240-244. https://doi.org/10.1016/j.procbio.2010.08.016
Sheela, S., Smita, V. and Dipak, V., 2016. Optimization of parameters for enhanced tannase production from a novel bacterial producer. World J. Pharm. Res., 5: 2131-2139.
Singh, S.K., Singh, S.K., Tripathi, V.R., Khare, S.K. and Garg, S.K., 2011. Comparative one-factor-at-a-time, response surface (statistical) and bench-scale bioreactor level optimization of thermoalkaline protease production from a psychrotrophic Pseudomonas putida SKG-1 isolate. Microb. Cell Fact., 10: 114. https://doi.org/10.1186/1475-2859-10-114
Sivashanmugam, K. and Jayaraman, G., 2011. Production and partial purification of extracellular tannase by Klebsiella pneumoniae MTCC 7162 isolated from tannery effluent. Afr. J. Biotechnol., 10: 1364-1374.
Souza, L.O., de Brito, A.R., Bonomo, R.C.F., Santana, N.B., Ferraz, J.L.A.A., Aguiar-Oliveira, E., Fernandes, A.G.A., Ferreira, M.L.O., de Oliveira, J.R. and Franco, M., 2018. Comparison of the biochemical properties between the xylanases of Thermomyces lanuginosus (Sigma®) and excreted by Penicillium roqueforti ATCC 10110 during the solid state fermentation of sugarcane bagasse. Biocatal. Agric. Biotechnol., 16: 277–284. https://doi.org/10.1016/j.bcab.2018.08.016
Talukdar, S., Ringø, E. and Ghosh, K., 2016. Extracellular tannase-producing bacteria detected in the digestive tracts of freshwater fishes (Actinopterygii: Cyprinidae and Cichlidae). https://doi.org/10.3750/AIP2016.46.3.04
Tripathi, A.D. and Lakshmi, B., 2018. Statistical optimization of extracellular tannase production by Streptomyces sp. AT 13 using response surface methodology and Plackett-Burmen design. Biosci. Biotechnol. Res. Commun., 11: 691-698. https://doi.org/10.21786/bbrc/11.4/21
Wang, F., Ni, H., Cai, H.N. and Xiao, A.F., 2013. Tea stalks–a novel agro-residue for the production of tannase under solid state fermentation by Aspergillus niger JMU-TS528. Annls Microbiol., 63: 897-904. https://doi.org/10.1007/s13213-012-0541-5
Wu, C., Zhang, F., Li, L., Jiang, Z., Ni, H. and Xiao, A., 2018. Novel optimization strategy for tannase production through a modified solid-state fermentation system. Biotechnol. Biofuels, 11: 92. https://doi.org/10.1186/s13068-018-1093-0
Xiao, A., Huang, Y., Ni, H., Cai, H. and Yang, Q., 2015. Statistical optimization for tannase production by Aspergillus tubingensis in solid-state fermentation using tea stalks. Electron. J. Biotechnol., 18: 143-147. https://doi.org/10.1016/j.ejbt.2015.02.001
Yee, T.W., Prabhu, N.G., Jain, K. and Ibrahim, D., 2011. Process parameters influencing tannase production by Aspergillus niger using mangrove (Rhizophora apiculata) bark in solid substrate fermentation. Afr. J. Biotechnol., 10: 13147-13154.
To share on other social networks, click on any share button. What are these?









