Isolation of Escherichia Coli Phages from Waste Waters
Research Article
Isolation of Escherichia Coli Phages from Waste Waters
Stephen Chijioke Emencheta1, Anthony Amaechi Attama2, Ezinwanne Nneoma Ezeibe1, Adaora Angela Agubata1 and Ebele Benedette Onuigbo1*
1Department of Pharmaceutical Microbiology and Biotechnology, Faculty of Pharmaceutical Sciences, University of Nigeria, Nsukka, 410001, Nsukka, Enugu State, Nigeria; 2Department of Pharmaceutics, Faculty of Pharmaceutical Sciences, University of Nigeria, Nsukka, 410001, Nsukka, Enugu State, Nigeria.
Abstract | This research was designed to isolate and test the lytic action of a bacteriophage specific to Escherichia coli. Both Escherichia coli and bacteriophages were isolated from farmlands and residential wastewaters. The isolated Escherichia coli were confirmed through phenotypic and biochemical tests. Antimicrobial susceptibility test to nine antibiotics was determined. Multi-antibiotic resistance index (MARI) was assessed. Spot assay was done to determine lytic action of the bacteriophage on the lawns of the Escherichia coli. The phenotypic and biochemical tests confirmed Escherichia coli isolates. There was metallic green sheen on EMB agar. The isolates were citrate negative and indole positive. The isolates were found to be resistant to amoxicillin (100 %), meropenem (100 %), and ceftriaxone (100 %). The Multi Antibiotic resistance index (MARI) of the isolates was calculated to be 0.33. The formation of plaques (clear zones of inhibition) on the lawn of the plates of Escherichia coli was confirmed. The isolation of Escherichia coli phages from residential and farm wastewaters is a promising molecular tool for E. coli tracking in the environment.
Received | February 17, 2022; Accepted | April 02, 2022; Published | April 27, 2022
*Correspondence | Ebele Benedette Onuigbo, Department of Pharmaceutical Microbiology and Biotechnology, Faculty of Pharmaceutical Sciences, University of Nigeria, Nsukka, 410001, Nsukka, Enugu State, Nigeria; Email: ebele.onuigbo@unn.edu.ng
Citation | Emencheta, S.C., Attama, A.A., Ezeibe, E.N., Agubata, A.A. and Onuigbo, E.B., 2022. Isolation of Escherichia coli phages from waste waters. Hosts and Viruses, 9: 12-18.
DOI | https://dx.doi.org/10.17582/journal.hv/2022/9.2.1.7
Keywords: Bacteriophage, Escherichia coli, Plaques, Lytic, Tracking, Antibiotic
Copyright: 2022 by the authors. Licensee ResearchersLinks Ltd, England, UK.
This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Introduction
Enterobacteriaceae are a large and diverse family of gram-negative non-spore forming bacilli, some of which are normal flora and some are pathogenic. Escherichia coli is a member of Enterobacteriaceae that can either be a commensal or cause intestinal or extraintestinal infections, such as diarrhoea, bleeding colitis, hemolytic uremic syndrome, thrombocytopenic purpura or death (Zared et al., 2021). Escherichia coli are one of the most studied bacteria in the world. This rod-shaped organism is normally discharged in the environment through open defecation of humans and animals or through waste-water treatment plants. Under optimal conditions, it can replicate in 20 minutes making it an ecological threat (Jang et al., 2017). There are six intestinal pathotypes causing a variety of human diseases and leading to an average of two million deaths a year. They are enterotoxigenic E. coli (ETEC), enteropathogenic E. coli (EPEC), shiga toxin-producing E. coli (STEC), enteroaggregative E. coli (EAEC), diffusely adherent E. coli (DAEC) and enteroinvasive E. coli (EIEC) (Jang et al., 2017). In recent times, metagenetics has been studying the active genetic exchange from a mixed community of organisms in the natural environment inducing the spread of virulence and resistance determinants (Klassert et al., 2021). Among the different antibiotic-resistant mechanisms developed by ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumonia, Acinetobacter baumanii, Pseudomonas aeruginosa, Enterobacteriaceae) pathogens, the ones found by Enterobacteriaceae are more diverse. Multidrug-resistant E. coli have become a global threat due to its ease of transmission among humans and from animals to humans through the faecal-oral route. E. coli has a dual functionality of acting as a donor of genetic material or recipient of resistant genes from other microorganisms (Galindo-Mendez, 2020). In the current era of increasing antibiotic resistance and lack of new antimicrobials, the re-emergence of bacteriophages becomes attractive. Bacteriophages are the most prolific biological form on Earth specific for eliminating bacteria. Each bacterial species is preyed by one or multiple bacteriophages which makes them a promising alternative for pathogenic control and ecological stability. This selectivity of bacteriophages is leveraged by environmental microbiologists in microbial source-tracking or monitoring (Rogovski et al., 2021; Attama et al., 2017). They are being used as indicators in fecal contamination (Poluri et al., 2021). This study aims to isolate coliphages from the community for further characterization as a molecular tool in environmental tracking of E. coli.
Materials and Methods
Study area and sample collection
The study was conducted in Nsukka, Enugu State, Nigeria. Samples of waste water were taken from ninety-two farms and residential areas over a period of three weeks. A 10 ml sample was collected in sterilized Bijou bottles and taken to the research laboratory for bacteriological analysis. Samples were randomly collected.
Isolation of Escherichia coli from the wastewater
The wastewater samples were diluted 10-fold with distilled water. A 0.1 ml of the diluted samples was dropped at the centre of different Petri dishes containing MacConkey agar. A spreader was used to spread the sample on the surface under aseptic conditions. The inoculated plates were incubated at 37 oC for 24 h in an autoclave.
Purification of isolated Escherichia coli
The overnight culture was purified by picking a colony from each plate and sub-culturing using the streak plate method on MacConkey agar and incubating for 24 h at 37 oC. Distinct colonies from each colony were then sub-cultured on Eosin Methylene blue (EMB) agar and incubated for 24 h at 37 oC to identify the Escherichia coli. Pure E. coli isolates were then stocked in double strength nutrient agar pending further tests.
Biochemical tests
Citrate and indole tests were carried out on 24 h broth cultures to confirm the presence of Escherichia coli.
Antimicrobial susceptibility testing
The antimicrobial susceptibility of E. coli isolates to aztreonam (ATM), chloramphenicol (CPL), gentamicin (GTN), tetracycline (TTE), sulphamethoxazole/trimethoprim (SXT), amoxicillin/clavulanic acid (AMC), meropenem (MEM), ciprofloxacin (CIP), ceftriaxone (CRO) was determined by the Kirby Bauer disk diffusion method according to CLSI guidelines (CLSI, 2014).
Multiple antibiotic resistance index (MARI)
Multiple Antibiotic Resistance Index (MARI) was determined using the procedure as described by Krumperman (1983). A MARI for an isolate was calculated as:
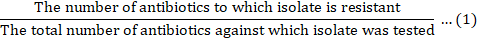
Isolation of bacteriophages
The E. coli phages were isolated from the farm or residential wastewater by centrifuging at 15,000 rpm for 10 min to separate the phages from other particles. The supernatant was filtered through 0.45µm syringe filter units. The filtrates were stored in the freezer.
Phage enrichment and filtration
The pure stocked isolates of E. coli were inoculated into 10 ml nutrient broth and incubated overnight. For the enrichment, the reaction mixture in a sterile test tube consisted of 0.4 ml of the overnight broth culture, 0.5 ml of 5x Luria Bertani (LB) broth, 0.04 ml calcium chloride (CaCl2), and 1.2 ml of the phage filtrate. This was incubated for 48 h at 37 oC. After incubation, the mixture was centrifuged for 15 min at 15,000 rpm. The supernatant was filtered using 0.45µm syringe filter units and the filtrates were stored at 4 oC.
Spot assay
A suspension of the 24 h bacterial culture, adjusted to 0.5 McFarland was swabbed on the surface of Mueller-Hinton agar plates and allowed to incubate at room temperature for 15 min. A drop of the enriched phages was spotted on the lawn of the bacterial isolate and allowed to incubate at room temperature for 15 min. The plates were incubated at 37 oC overnight. After incubation, the plates were observed for the presence of plaques.
Results and Discussion
Isolation and biochemical tests
From a total of ninety-two (92) isolates collected from different locations of waste waters, twenty-one (21) isolates were suspected to be E. coli because of the pink appearance on MacConkey agar. These 21 isolates were then subcultured on EMB agar and only 8 isolates had the distinctive metallic green sheen which is 8.69 % of the total samples collected. All eight isolates were subjected to biochemical tests and the results are shown in Table 1. A negative reaction was obtained with the citrate utilization test while a positive reaction was detected in the indole test. Identification was based specifically on the positive indole reaction. It was thought that E. coli cannot survive outside the intestinal tract for long periods of time but recent studies have shown that this bacterium can naturalize and reproduce in soil, sand and sediments in tropical, subtropical and temperate climates (Byappanahalli et al., 2015). Contamination of drinking water by this waste water through surface run-offs or flooding could cause illnesses such as urinary tract infection, meningitis, septicaemia or intestinal infections when harbouring toxins from the E. coli. Adapted E. coli in the environment have been detected with high levels of eaeA, which encodes intimin, a virulence factor. Presence of eaeA in the environment is a great concern for public health. Escherichia coli O157: H7 have also been known to persist for months in manure and on green leafy vegetables such as spinach and lettuce and cause outbreaks (Ishii et al., 2014). Some ready-to-eat foods and vegetables grown on and irrigated with these waste waters get contaminated with E. coli at different stages from pre- to postharvest (Luna-Guevara et al., 2019). Boiling of drinking water at 100 oC for some minutes, handling and good storage of water becomes important in households as a preventive measure (WHO, 2017). Basic hygiene and handling of foods for personnel at farms, abbatoirs and restaurants is equally essential to keep E. coli infection to a minimum (WHO, 2017).
Table 1: Biochemical tests to confirm presence of E. coli in the environment.
|
S/n |
Isolate |
Source |
||
|
Citrate |
Indole |
|||
|
1 |
42 |
Res |
- |
+ |
|
2 |
48 |
Res |
- |
+ |
|
3 |
78 |
Farm |
- |
+ |
|
4 |
82 |
Farm |
- |
+ |
|
5 |
83 |
Res |
- |
+ |
|
6 |
85 |
Res |
- |
+ |
|
7 |
89 |
Farm |
- |
+ |
|
8 |
90 |
Res |
- |
+ |
Key: Res(Residential), +(positive), - (negative).
Antimicrobial susceptibility testing
The isolated strains of E. coli were screened against aztreonam (ATM), chloramphenicol(CPL), gentamicin(GTN), tetracycline (TTE), amoxicillin/ clavulanic acid (AMC), sulphamethoxazole/ trimethoprim (SXT), ciprofloxacin (CIP), meropenem (MEM), and ceftriaxone (CRO). There was high susceptibility of the strains to ciprofloxacin (CIP), followed by sulphamethoxazole/ trimethoprim and chloramphenicol respectively as shown in Table 2. However, there was 100 percent resistance to amoxicillin/ clavulanic acid, meropenem and ceftriaxone. The role of environmental factors in the antibiotic resistance transmission and migration dynamics of Escherichia coli is not well understood. However, the ecological consequences of the antimicrobial resistance are difficult to control because of close interactions between interfacing environments (Guangshui et al., 2018). The increased antimicrobial resistance in the environment could be attributed to increased anthropogenic activities, some inappropriate, involving antimicrobial compounds and subsequent release into receiving environments such as soil and water. Researchers are undertaking studies on the chemicals and pollutants being released into the environment and the pressure of resistant pathogens being selected. It behoves policy-makers to study these findings and take appropriate actions to control the unwanted effects (Stanton et al., 2020). E. coli has evolved different mechanisms of resistance to antibiotics mostly plasmidic and which transfers
Table 2: Antimicrobial susceptibility test of Escherichia coli to antibiotics of choice.
|
S/n |
Isolate |
Source |
ATM 30µm |
CPL 30µm |
GTN 10µm |
TET 30µm |
SXT 25µm |
AMC 30µm |
MEM 10µm |
CIP 5µm |
CRO 30µm |
|
|
1 |
42 |
Res |
15 I |
16 S |
9 I |
12 I |
18 S |
0 R |
0 R |
29 S |
0 R |
|
|
2 |
48 |
Res |
13 I |
18 S |
6 I |
10 I |
20 S |
0 R |
0 R |
30 S |
0 R |
|
|
3 |
78 |
farm |
11 I |
16 S |
7 I |
9 I |
17 S |
0 R |
0 R |
28 S |
0 R |
|
|
4 |
82 |
farm |
12 I |
16 S |
9 I |
8 I |
19 S |
0 R |
0 R |
26 S |
0 R |
|
|
5 |
83 |
Res |
13 I |
19 S |
10 I |
11 I |
16 S |
0 R |
0 R |
29 S |
0 R |
|
|
6 |
85 |
Res |
14 I |
23 S |
13 I |
10 I |
19 S |
0 R |
0 R |
34 S |
0 R |
|
|
7 |
89 |
Farm |
10 I |
17 S |
13 I |
7 I |
16 S |
0 R |
0 R |
34 S |
0 R |
|
|
8 |
90 |
Res |
7 I |
20 S |
6 I |
13 I |
17 S |
0 R |
0 R |
30 S |
0 R |
|
Key: S-susceptible; I-Intermediate; R-Resistant.
resistance genes encoding extended spectrum β-lactamases (ESBLs) horizontally to other species including pathogenic bacteria (Peterson and Kauer, 2018). The most commonly seen ESBLs in E. coli are CTX-M enzymes. There is now an emergence of carbapenemase-producing enterobacteriaceae against carbapenem which has been used as the first-line drug for ESBLs. Current carbapenemases are Ambler Class A Klebsiella pneumonia carbapenemase (KPC); Class B metallo-β-lactamases (MBLs) such as New Delhi MBL (NDM), Verona integrin-encoded MBL (VIM), and imipenemase (IMP); and Class D oxacillinases (OXA)-type enzymes such as OXA-48-like carbapenemases. A novel boron-containing serine-β-lactamase inhibitor called Varborbactam is being studied which has inhibitory activity against Ambler Class A and C serine carbapenemases such as KPC. It is used synergistically in combination with meropenem as a carbapenem-β-lacatamase inhibitor with susceptibility rates ranging from 66.2 to 100 % (Sheu et al., 2019). World Health Organization (WHO) announced that one of the top ten global health threats is Antimicrobial resistance (AMR) (WHO, 2020). It has been forewarned that by, 2050, antibiotic resistance will be responsible for the death of 10 million people (Sharma et al., 2021). In another study, AMR was projected to drain the global economy of US$100 trillion in the same period (Mogasale et al., 2021). From the antibiotic susceptibility study, Escherichia coli exhibited moderate to high resistance pattern with a MARI of 0.3. MARI is a dependable and reproducible method of tracking antibiotic resistance in the environment. The major factors causing the high resistance of bacteria to multiple antibiotics is the indiscriminate use of antibiotics in homes and farms (Ayandele et al., 2020).
Table 3: Showing the number of plaques formed by the bacteriophages.
|
S/N |
Isolates |
Code |
Source |
Phage filtrates |
|||
|
Spots/Plaques |
|||||||
|
1 |
2 |
3 |
4 |
||||
|
1 |
42 |
R |
RES |
+ |
- |
- |
+ |
|
2 |
85 |
S |
Sand |
+ |
- |
- |
+ |
Spot assay
The Muller-Hinton agar plates containing lawns of eight confirmed E. coli isolates were spotted with the enriched bacteriophages in four different spots. After 24 h incubation, we observed only two plates with plaques as seen in Table 3. This test was to done to determine the susceptibility of Escherichia coli to a lytic concentration of a specific bacteriophage(s). The spot test revealed that the enriched phages were capable of infecting some of the isolated Escherichia coli from different environmental waste waters. The phages showed clear plaques and no overgrowth was observed. An increasing number of studies have examined the use of phages in Escherichia coli tracking or as indicators of fecal contamination in an environment. This has been done through phage-induced bacterial lysis using quantitative image analysis to achieve detection of Escherichia coli at low concentrations (Yang et al., 2020). An imaging study using rhodamine isothiocyanate or DNA dye Syto 13 fluorescently labeled phages and its Escherichia coli receptor, LamB have also been studied to reveal the role of space and time in the evolutionary dynamics of phage control with high throughput outcomes (Wang et al., 2020; Low et al., 2020). A study has also investigated the use of transposon insertion sequencing (INSeq) screens as a rapid, high throughput tool to identify candidate phage receptors (bacterial genes involved in phage binding) for previously well characterized phages T2, T4, T6 and T7 (Kortright et al., 2020). In Bangkok canals, the microbial quality was assessed using the fluorescent bacteriophage assay (FBA) and fluorescence in-situ hybridization (FISH) using oligonucleotide probes targeted to Escherichia coli (Takehiko et al., 2006).
Conclusions and Recommendations
The phages isolated from farms and residential waste waters were specific to the Escherichia coli isolates. The re-emergence of phages as an environmental tool for Escherichia coli tracking is both an iterative and attractive development for ecologists. Further studies would be done to characterize the phages isolated.
Novelty Statement
The study contributes to the increasing support in the future advancement and utilization of bacteriophages in common biological processes. Specifically here, the use of specific bacteriophages as a tool for identification, monitoring, and control of specific Escherichia coli strains in the environment.
Author’s Contribution
The study was conceptualized by Ebele Benedette Onuigbo and Anthony Amaechi Attama. Experimentation and data collection was done by Stephen Chijioke Emencheta and Adaora Angela Agubata. Initial manuscript was prepared by Ebele Benedette Onuigbo and Stephen Chijioke Emencheta. Manuscript was reviewed for intellectual content by Ezinwanne Nneoma Ezeibe and Anthony Amaechi Attama.
Conflict of interest
The authors have declared no conflict of interest.
References
Attama, A.A., Agbo, I.S., Eke, I.E., Onuigbo, E.B., and Ogbonna, J.C., 2017. Bacteriophage: Clinical applications. In: Mendez-Vilas, A (ed), Antimicrobial research: Novel bioknowledge and educational programs, 1st ed pp. 260-269.
Ayandele, A.A., Oladipo, E.K., Oyebisi, O., and Kaka, M.O., 2020. Prevalence of Multi-Antibiotic Resistant Escherichia coli and Klebsiella species obtained from a tertiary medical institution in Oyo State, Nigeria. Qatar Med. J., 1: 9. https://doi.org/10.5339/qmj.2020.9
Byappanahalli, M.N., Nevers, M.B., Whitman, R.L., and Ishii, S., 2015. Application of a microfluidic quantitative polymerase chain reaction technique to monitor bacterial pathogens in beach water and complex environmental matrices. Environ. Sci. Technol. Lett., 2: 347-351. https://doi.org/10.1021/acs.estlett.5b00251
Cag, Y., Caskurlu, H., Fan, Y., Cao, B., and Vahaboglu, H., 2016. Resistance mechanisms. Ann. Transl. Med., 4(17): 326. https://doi.org/10.21037/atm.2016.09.14
Clinical and Laboratory Standards Institute, 2014. Performance standards for antimicrobial susceptibility testing 24th ed, CLSI Supplement M100-S24, Wayne, PA, USA, pp. 50.
Diggle, S., and Whiteley, M., 2020. Microbe Profile: Pseudomonas aeruginosa: Opportunistic pathogen and lab rat. Microbiology, 166(1): 30-33.
Driesch, D., Gasteneier, P., and Slevogt, H., 2021. Bacterial colonization dynamics and antibiotic resistance gene dissemination in the hospital environment after first patient occupancy: A longitudinal metagenetic study. BMC Microbiome, 9(169): https://doi.org/10.1186/s40168-021-01109-7
Galindo-Mendez, M., 2020. Antimicrobial resistance in Escherichia coli. In: Luis Rodrigo (Eds.) E. coli Infections-Importance of early diagnosis and efficient treatment. IntechOp., 1-21. https://doi.org/10.5772/intechopen.93115
Guangshui, N., Zihao, L., Hui, G., Linxiaozhang, Qianwei, L., Ruijing L., Fan, Y., Chanlin, H. and Ziwei, Y., 2018. The effect of environmental factors and migration dynamics on the prevalence of antibiotic-resistant Escherichia coli in estuary environments. Scient. Rep., 8: 1663. https://doi.org/10.1038/s41598-018-20077-x
Ishii, S., Nakamura, T., Ozawa, S., Kobayashi, A., Sano, D., and Okabe, S., 2014. Water quality monitoring and risk assessment by simultaneous multipathogen quantification. Environ. Sci. Technol. 48: 4744-4749. https://doi.org/10.1021/es500578s
Jang, J., Hur, H.G., Sadowsky, M.J., Byappanahalli, M.N., Yan, T., and Ishili, S., 2017. Environmental Escherichia coli: Ecology and public health implications. A review. J. Appl. Microbiol., 123(3): 570-581. https://doi.org/10.1111/jam.13468
Klassert, T.E., Leistner, R., Zubiria-Barrera, C., Stock, M., Lopez, M., Neubert, R., Driesch, D., Gasteneier, P. and Slevogt, H., 2021. Bacterial colonization dynamics and antibiotic resistance gene dissemination in the hospital environment after first patient occupancy: a longitudinal metagenetic study. Microbiol., 9(169): https://doi.org/10.1186/s40168-021-01109-7
Kortright, K.E., Chan, B.K., and Turner, P.E., 2020. High-throughput discovery of phage receptors using transposon insertion sequencing of bacteria. Proc. Natl. Acad. Sci., 117(31): 18670-18679. https://doi.org/10.1073/pnas.2001888117
Krumperman, P.H., 1983. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl. Environ. Microbiol., 46: 165-170.
Low, H.Z., Bohnlein, C., Sprotte, S., Wagner, N., Fiedler, G., Kabisch, J., and Franz, C., 2020. Fast and easy phage-tagging and live/ dead analysis for the rapid monitoring of bacteriophage infection. Front. Microbiol., 11(602444): 1-13.. https://doi.org/10.3389/fmicb.2020.602444
Luna-Guevara, J.J., Arenas-Hernadez, M.M.P., Martinez de la Pena, C., Silva, J.L., and Luna-Guevara, M.L., 2019. The role of pathogenic E. coli in fresh vegetables: Behaviour, contamination factors, and preventive measures. Int. J. Microbiol., Volume 2019, Article ID 2894328. https://doi.org/10.1155/2019/2894328
Mogasale, V., Saldanha, P., and Mogasale, V., 2021. A descriptive analysis of antimicrobial resistance patterns of WHO priority pathogens isolated in children from a tertiary care hospital in India. Sci. Rep., 11: 5116. https://doi.org/10.1038/s41598-021-84293-8
Na, G., Lu, Z., Gao, H., Li, Q., Li, R., Yang, F., Huo, C., and Yao, Z., 2018. The effect of environmental factors and migration dynamics on the prevalence of antibiotic-resistant Escherichia coli in estuary environments. Sci. Rep., 8: 1663. https://doi.org/10.1038/s41598-018-20077-x
Peterson, E., and Kaur, P., 2018. Antibiotic resistance mechanisms in Bacteria: Relationships between resistance determinants of antibiotic producers, environmental bacteria, and clinical pathogens. Front. Microbiol., 9(2928): 1-21. https://doi.org/10.3389/fmicb.2018.02928
Poluri, K.M., Sitthisak, S., Khairnar, K., and Czajkowski, R., 2021. Editorial: Bacteriophages isolation from the environment and their antimicrobial therapeutic potential. Front. Microbiol., 12(649334): 1-4. https://doi.org/10.3389/fmicb.2021.649334
Rogovski, P., Cadamuro, R.D., da Silva, R., Brasiliense de Souva, E., Bonatto, C., Viancelli, A., Michelon, W., Elmahdy, E.M., Treichel, H., Rodriguez-Lazaro, D., and Fongaro, G., 2021. Uses of bacteriophages as bacterial control tools and environmental safety indicators. Front. Microbiol., 12(793135): 1-11. https://doi.org/10.3389/fmicb.2021.793135
Sharma, S., Datta, S., and Dwivedi, S.K., 2021. Isolation and characterization of a lytic bacteriophage against Pseudomonas aeruginosa. Sci. Rep., 11: 193093. https://doi.org/10.1038/s41598-021-98457-z
Sheu, C., Chang, Y., Lin, S., Chen, Y., and Hsueh, P., 2019. Infections caused by Carbapenem-resistant Enterobacteriaceae: An update on therapeutic options. Front. Microbiol., 10(80): 1-13. https://doi.org/10.3389/fmicb.2019.00080
Stanton, I.C., Bethel, A., Leonard, A.F.C., Gaze, W.H., and Garside, R., 2020. What is the research evidence for antibiotic resistance exposure and transmission to humans from the environment? A systematic map protocol. Environ. Evid., 9: 12. https://doi.org/10.1186/s13750-020-00197-6
Takehiko, K., Fuangfa, U., Orasa, S., and Masao, N., 2006. Rapid monitoring of Escherichia coli in Southeast Asian Urban canals by fluorescent-bacteriophage assay. J. Health Sci., 52: 666-671. https://doi.org/10.1248/jhs.52.666.
Tilman, E., Klassert, R.L., Zubiria-Barrera, C., Stock, M., Lopez, M., Neubert, R., and Galindo-Mendez, M., 2020. Antimicrobial resistance in Escherichia coli. In: Luis Rodrigo (ed.) E. coli Infections-Importance of early diagnosis and efficient treatment. https://doi.org/10.5772/intechopen.93115
Wang, J., Applegate, B., Kanach, A., and Han, R., 2020. Applications of bacteriophage in rapid detection of Escherichia coli in foods. Curr. Opin. Food Sci., 39: 43-50. https://doi.org/10.1016/j.cofs.2020.12.015
World health Organization, (2017). Guideline for drinking-water quality: fourth edition incorporating the addendum. WHO Library Cataloguing-in-Publication Data. Geneva.
World Health Organization and Food and Agriculture Organization of the United Nations, 2020. Shiga toxin-producing Escherichia coli (STEC) and food: Attribution, characterization, and monitoring: Report. World Health Organization, 2020; https://apps.who.int/iris/handle/10665/272871
Yang, X., Wisuthiphaet, N., Young, G.M., and Nitin, N., 2020. Rapid detection of Escherichia coli using bacteriophage-induced lysis and imaging analysis. PLoS One, 15(6): e0233853. https://doi.org/10.1371/journal.pone.0233853
Zarei, O., Shokoohizadeh, L., Hossainpour, H., and Alikhani, M.Y., 2021. The prevalence of Shiga-Toxin producing Escherichia coli and Enteropathogenic Escherichia coli isolated from raw chicken meat samples. Int. J. Microbiol., Volume 2021, Article ID 3333240. https://doi.org/10.1155/2021/3333240
To share on other social networks, click on any share button. What are these?






