In-Silico Development of 1, 7-Dihydrodipyrrolo [2,3-b:3’,2’-e] Pyridine -3-carboxamide Derivatives as Candidate Janus Kinase Inhibitors to Control Rheumatoid Arthritis
In-Silico Development of 1, 7-Dihydrodipyrrolo [2,3-b:3’,2’-e] Pyridine -3-carboxamide Derivatives as Candidate Janus Kinase Inhibitors to Control Rheumatoid Arthritis
Neelam Pery*, Nayab Batool Rizvi, Muhammad Imtiaz Shafiq
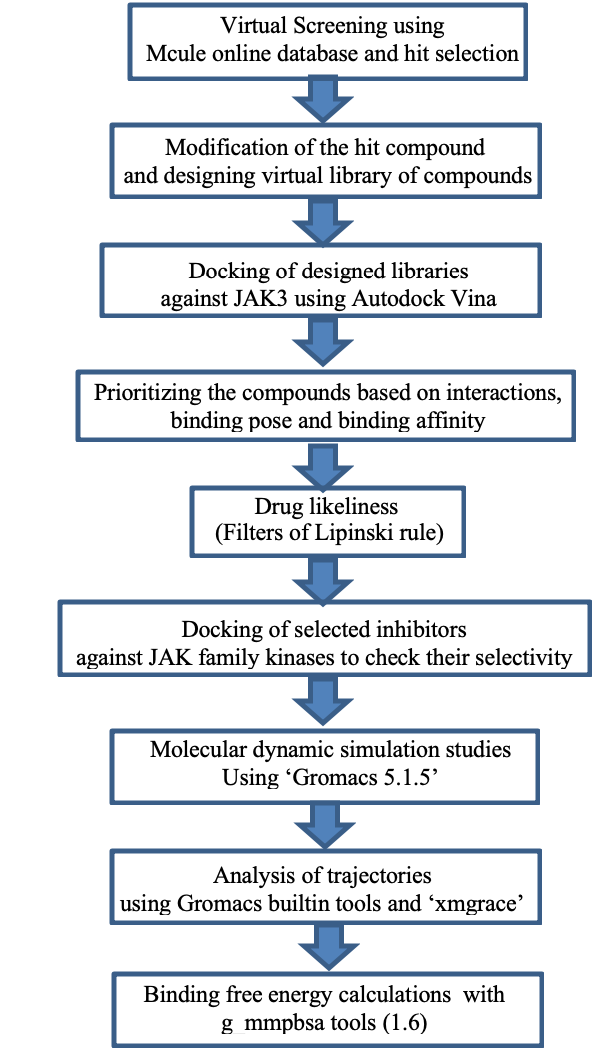
A schematic representation of the overall workflow used for drug designing.
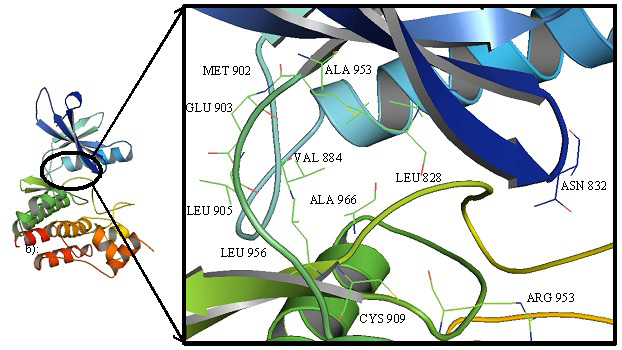
Cartoon representation of JAK3 highlighting the interacting residues in the binding cavity.
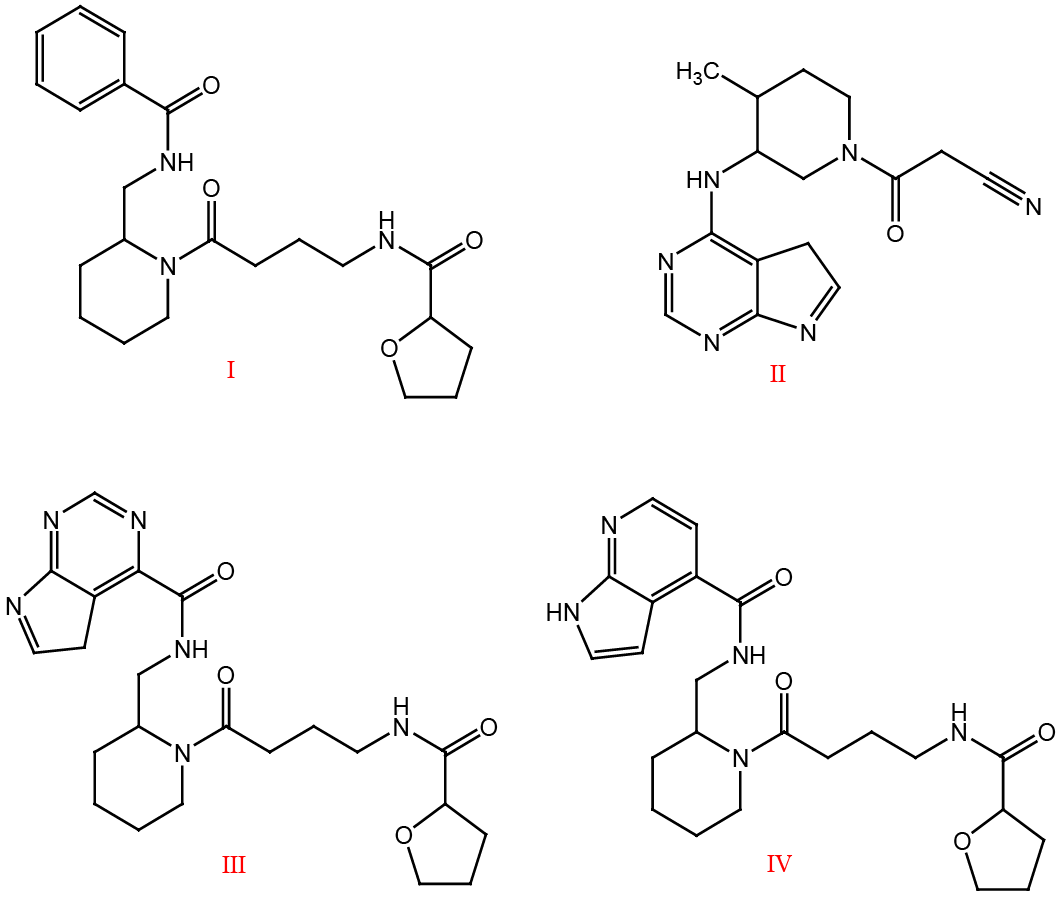
Virtual screening hit (Compound I), Tofacitinib (Compound II), Designed hybrids (III and IV).
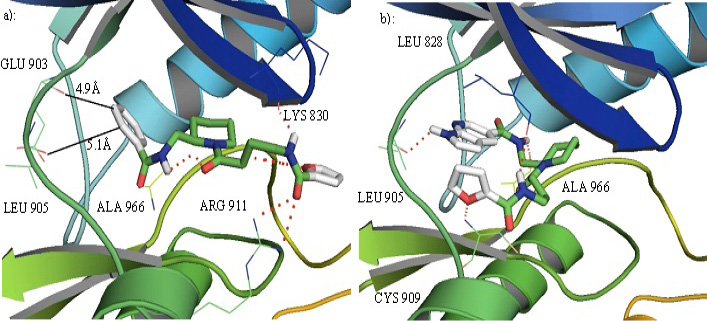
a): compound I in the binding pocket of JAK3. It can develop favorable interactions with different residues in the binding cavity but does not develop any interactions with the hinge residues GLU 903 and LEU 905. b): Compound IV in the binding pocket of JAK3 can interact with the hinge ATP binding residue and some other residues in the binding cavity.
Different fragments used to substitute the hinge binding fragment of compound IV.
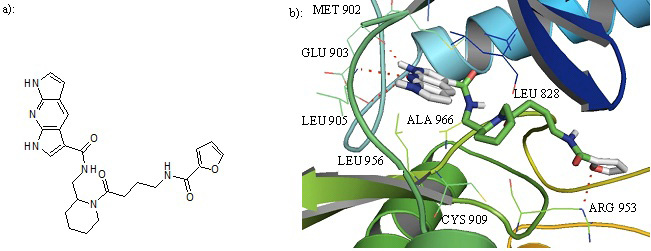
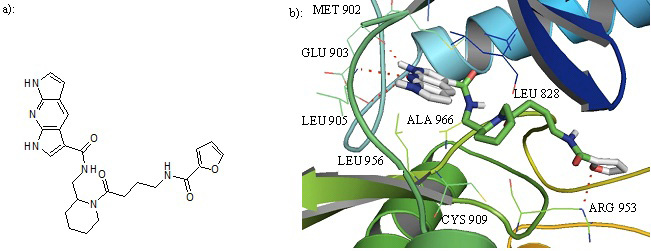
a): Structure of compound 27. b): Cartoon representation of Compound 27 in the binding pocket of JAK3. Dipyrrolopyridine moiety develops hydrogen bonds to the GLU 903 and LEU 905 of the hinge region while its furanamide moiety interacts with ARG 953 from the lower lobe.
Different fragments used to replace the piperidine ring of the designed inhibitor.
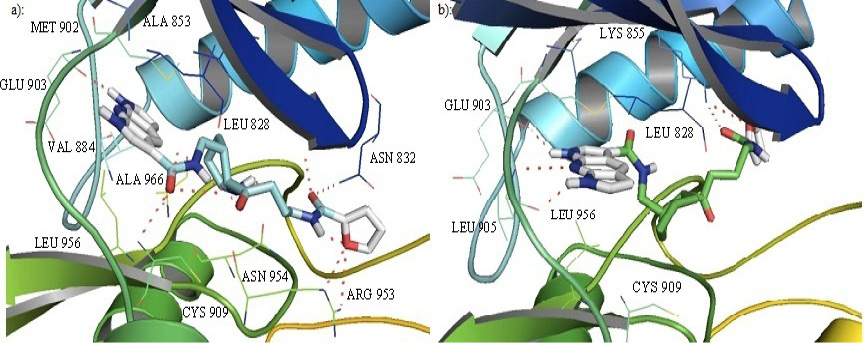
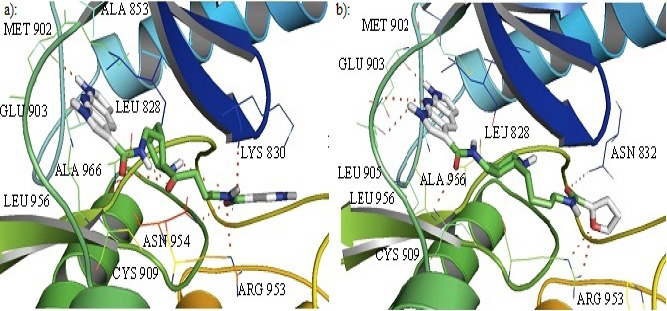
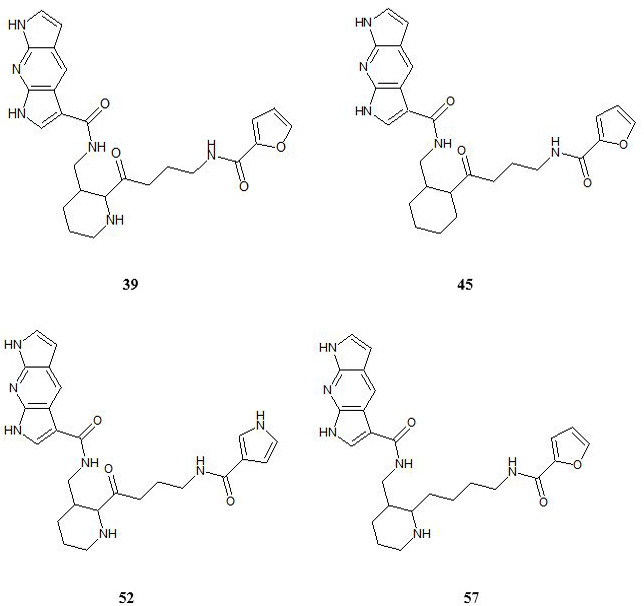
Cartoon representation of the binding pocket of JAK3 with compound 39 (a) and 45 (b). In 39, dipyrrolopyridine moiety develops hydrogen bonds to the GLU 903 and LEU 905 of the hinge region while its furanamide moiety interacts with ARG 953 from the lower lobe and ASN 832 from the upper lobe. In compound 45, dipyrrolopyridine moiety develops hydrogen bonds to the GLU 903 and LEU 905 hinge residue while its furanamide moiety makes a bidentate hydrogen bond to the LYS 855 and develops many other interactions with residues from the upper and lower lobe. Hydrogen bonding is represented by dotted lines. All the interacting residues are shown in the line model.
Substituents for the terminal furan ring of the designed inhibitors.
Different linker groups used for the designed inhibitors.
Cartoon representation of the binding pocket of JAK3 with inhibitor 52 (a) and 57(b). In inhibitor 52, dipyrrolopyridine moiety develops hydrogen bond to the GLU 903 while its furanamide moiety interacts with ARG 953 and ASN 954 from the lower lobe as well as LYS 830 from the upper lobe. The inhibitor also makes a hydrogen bond to CYS 909. b): Dipyrrolopyridine moiety develops three hydrogen bonds to the GLU 903 and LEU 905 of the hinge residues. Hydrogen bonding is also seen with ASN 832 from the upper lobe and ARG 953 and CYS 909 from the lower lobe. Hydrogen bonding is represented by dotted lines.
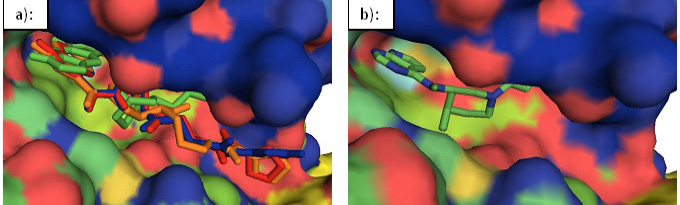
Surface representation of JAK3. a): An overlay of the designed inhibitors 39 (red), 45(green), 52 (blue), and 57 (orange) in the binding cavity of JAK3. b): Tofacitinib in the binding cavity of JAK3.
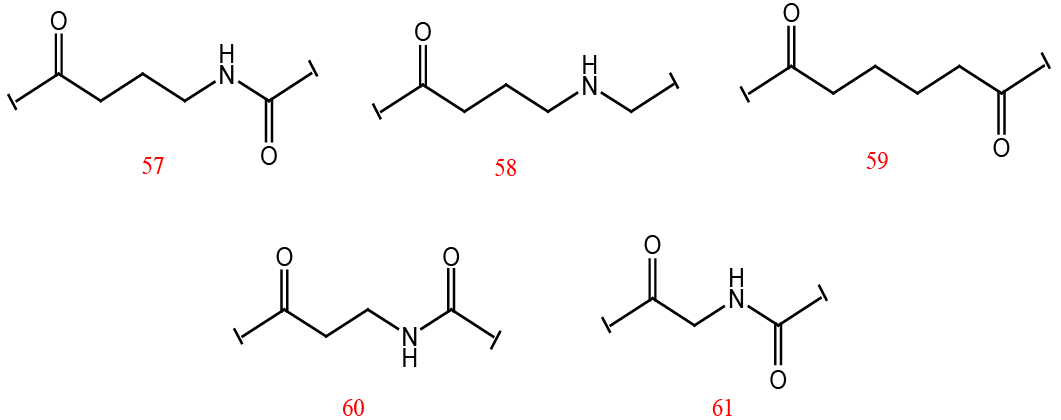
Structures of the designed inhibitors.
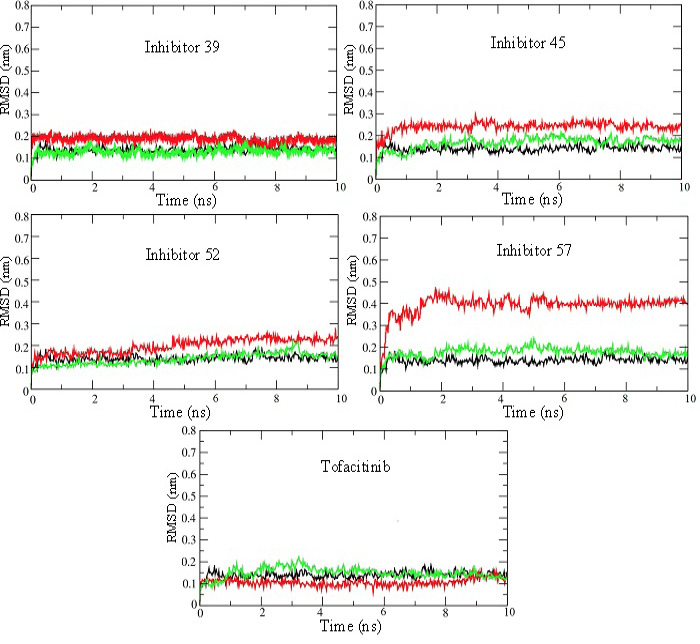
RMSD plots for 10 ns MD production run of simulations for JAK3 enzyme-inhibitor complexes. RMSD plots for the unbound enzyme (black), inhibitor (red), and enzyme bound with inhibitor (green) have been shown for selected inhibitors 39, 45, 52, 57, and Tofacitinib. These plots show that the systems attain stability very quickly and remain at steady-state throughout 10 ns simulations.
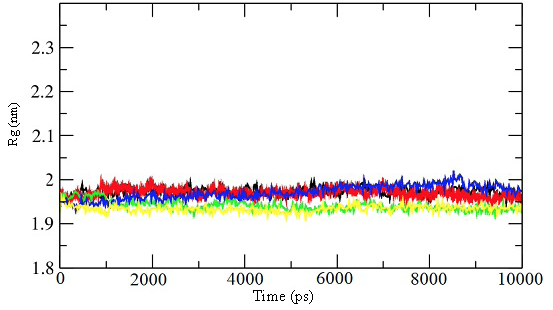
Radius of gyration for the 10 ns simulations for the unbound enzyme (black) and enzyme-inhibitor complex for inhibitor 39 (red), 45 (green), 52 (blue), and 57 (yellow).
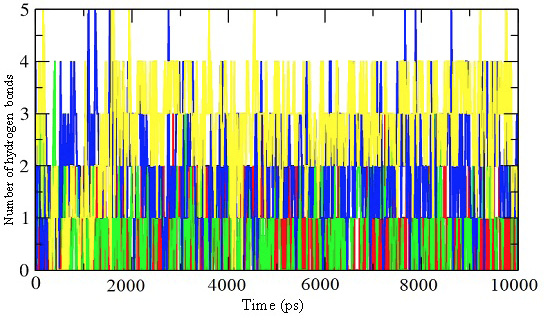
Representation of the number of hydrogen bonds per time frame for the enzyme-inhibitor complex of inhibitor 39 (red), 45 (green), 52 (blue), and 57 (yellow).
a): Structure of compound 27. b): Cartoon representation of Compound 27 in the binding pocket of JAK3. Dipyrrolopyridine moiety develops hydrogen bonds to the GLU 903 and LEU 905 of the hinge region while its furanamide moiety interacts with ARG 953 from the lower lobe.