Genomic Regions Associated with Early/Late Feathering in Daheng Broilers Identified Using Genome-Wide Association Studies
Genomic Regions Associated with Early/Late Feathering in Daheng Broilers Identified Using Genome-Wide Association Studies
Donghao Zhang1, Lingqian Yin1, Zhongzhen Lin1, Chaowu Yang2, Chunlin Yu1,2, Zengrong Zhang2, Mohan Qiu2* and Yiping Liu1*
1Farm Animal Genetic Resources Exploration and Innovation Key Laboratory of Sichuan Province, Sichuan Agricultural University, Chengdu, Sichuan 611130, China
2Animal Breeding and Genetics Key Laboratory of Sichuan Province, Sichuan Animal Science Academy, Chengdu, Sichuan 610066, China
ABSTRACT
A causal mutation of a repeat segment on the Z chromosome directly regulates the late feather phenotype in chickens. However, further research on the molecular mechanism of autosomal influence on feather development is of great significance for chicken production and early sex identification. Here, genotyping-by-sequencing (GBS) technology was used to develop genome-wide single nucleotide polymorphisms (SNPs) markers for early-feathering (EF) and late-feathering (LF) phenotypes in Daheng broilers. A total of 11626 high-quality SNPs were identified, and the different positions and numbers of SNPs were counted. On this basis, genome-wide association study (GWAS) analysis was performed to find molecular genetic markers significantly related to this trait. Candidate genes such as TBC1D22A, PRSS23 and HOXD gene families that may be related to feather growth and development were identified, and biological pathways to play their roles were sought. The concentration of these significant candidate genes in autosomal genes indicates that feathering rate is also related to autosomal inheritance. Then, each candidate SNPs and population frequency information were determined according to the sequencing results. Bayesian models were used to estimate the probability of LF or EF for each observed genotype. In all, our results improve the molecular regulatory mechanisms of follicle development and feathering phenotype, and provide data support for the establishment, refined production management and utilization of EF and LF lineages.
Article Information
Received 02 March 2023
Revised 26 March 2023
Accepted 09 April 2023
Available online 28 July 2023
(early access)
Published 08 January 2025
Authors’ Contribution
YPL and MHQ conceived the
research. DHZ, PR and JJL analyzed
the data. LQY, ZZL, CWY and CLY
conducted the experiments. DHZ wrote the manuscript.
Key words
Daheng broilers, Early-feathering, Late-feathering, Genome-wide association study, Bayesian model
DOI: https://dx.doi.org/10.17582/journal.pjz/20230302070341
* Corresponding author: liuyp578@163.com, mohan.qiu@163.com
0030-9923/2025/0001-0213 $ 9.00/00
Copyright 2025 by the authors. Licensee Zoological Society of Pakistan.
This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Introduction
EF and LF are sex-linked phenotypes, and one day of age is considered the best period for automatic sex determination (Zhao et al., 2016). The alleles (K-k+) are sex genes, located on the short arm of the chicken’s Z chromosome (Pal and Singh, 1997). The early and late feathers are controlled by a pair of alleles (K-k), and the K allele are incomplete dominant to k+ allele (Elferink et al., 2008). Among them, the K allele was related to LF, which retard the emergence of primary and secondary flight feathers, while the k+ allele resulting in the earliest emergence of feathers. Based on the length of primary feathers and primary-convert feathers at hatching, the EF phenotype is roughly divided into two types: more than 5mm and the difference between 2-5mm, while the classification and distribution of the LF phenotype are different. Some studies have divided it into five types, namely C (longer and isochron), D (shorter and isochron), E (large gap and isochron), F (small gap and isochron > G), but the distribution ratio has not been reported (Song et al., 2003). The establishment of early and late feathered strains by phenotypic selection or molecular identification is of great significance for chicken breeding and scientific management, which is reflected in the rapid operation, low cost and low chick stress response.
Although the LF/EF phenotype is beneficial for chick sexing, the relationship between the feather rate phenotype and performance is also being explored. Since plumage have different heat preservation ability at different temperatures, different feather types indirectly affect feed conversion rate and fat deposition, and thus affect carcass weight of poultry (Fotsa et al., 2001; Merkley and Lowe, 1988). LF/EF phenotype affects egg production and quality. Researches have shown that the egg production of brown layers at sexual maturity in LF group was lower than that in EF group (Collins et al., 1970). Compared with yellow-feathered broilers with LF phenotype, laying rate and hatching performance of EF phenotype were significantly increased (Goger et al., 2017). In addition, the lesion score indicated that the resistance of chicks to E. coli inoculation was associated with the K allele (Dunnington et al., 1986). Therefore, it is of great significance to study the mechanism of feather difference for sex identification and economic character improvement of chickens.
In recent years, sequencing technology has profoundly changed the way we understand genomes, enriching the genetic and biological inquiry into complex traits (Fuentes-Pardo and Ruzzante, 2017; Visscher et al., 2012). Based on the low cost of genotyping, high-throughput single nucleotide polymorphism (SNP) genotyping assays are increasingly used in genome-wide association studies in chickens and as a breeding tool (genome prediction and selection) (Groenen et al., 2011). GWAS is a genome-wide trait mapping method to locate genetic loci associated with traits by testing the significance of association between marker polymorphisms and phenotypic diversity of target traits (Sharma et al., 2015). In this context, Onteru et al. (2013) performed GWAS using a dense SNP panel to identify SNPs associated with FE-related traits and incorporated them into the prediction model (Onteru et al., 2013). Previous studies have identified two candidate genes associated with the chicken feather K allele: prolactin receptor (PRLR) (Bu et al., 2013) and sperm flagellum 2 (SPEF2) (Elferink et al., 2008). Both have been identified as biomarkers of chicken feathering. However, more candidate genes and more comprehensive identification methods for feathering traits need to be developed. In the actual production process, although the gender of chickens is mainly identified according to the development of wing feathers, but the success rate of identification is still worth improving.
In this study, genotyping-by-sequencing (GBS) technology was used to develop genome-wide SNP markers for early and late feathering traits in Daheng broilers (55 samples in group K and 54 samples in group M). On this basis, GWAS analysis was carried out to find out the molecular genetic markers significantly related to this trait, so as to provide theoretical support for later molecular breeding technology.
MATERIALS AND METHODS
Animal samples
All the chickens used in the experiments were provided by Sichuan Daheng Poultry Breeding Co. LTD, which were raised in a traditional environment, with the same temperature and humidity and with free access to water and feed.
Phenotypes of the feather rate in one-day-old chicks were distinguished according to Xie et al. (1985). Briefly, 1-day-old chicks with a primary feather more than 2 mm longer than the primary-covert feathers is an EF phenotype, otherwise defined as a LF phenotype, which can be divided into inverted, equal and slightly long types (Fig. 1). Then, blood samples were collected from a total of 99 1-day-old Daheng broilers which provided by Sichuan Daheng Poultry Breeding Co. LTD, including 55 EF phenotype chickens and 54 LF phenotype chickens, according to the method of Luo et al. (2012). In brief, chickens were sterilized with alcohol, followed by 1 ml venous blood drawn using a 2 ml medical syringe and stored immediately in a -20℃ refrigerator.
DNA library construction and quality control
The DNA concentration was accurately quantified by Qubit and concentrated to 50 ng/μL (Wang et al., 2017). The entire library was prepared using a Covaris ultrasonic crushing machine with random interruptions according to Illumina’s official procedures. Then, Qubit2.0 was used for initial quantification, the library was diluted to 1.5ng/μl, and the insert size of the library was detected using Agilent 2100. Illumina NovaSeq6000 sequencing platform was used for PE150 mode sequencing; the obtained Raw data were transformed into Raw sequencing sequences by base recognition, called Raw Reads, which were filtered for quality using Fastp software (Chen et al., 2018). The following filtering parameters were adopted: Remove the reads that (i) contained adaptor and (ii) more than 3 unknown bases (N bases) and (iii) those, in which one end had more than 20% low quality base reads (sequencing quality value< 5).
SNP genotyping
The BWA software (Li and Durbin, 2009) was used to compare Clean Reads to the chicken reference genome (GCF_016699485.2_bGalGal1.mat.broiler.GRCg7b_genomic.fa), and then SAM tools software (Li et al., 2009) was used to convert the initial alignment results into BAM format and sort them.
Variant detection and annotation
The haplotype caller model of GATK software (Li et al., 2009) was used to detect SNP and InDel in a single sample. Finally, the genotype GVCF files of individual samples were merged using the combine variants function of GATK software, and the genotype GVCFs function was used for population SNP detection. SNP quality filtering was performed using Vcf tools (Danecek et al., 2011) and Plink software (Purcell et al., 2007). The set was then filtered using the following criteria: (i) maximum alleles number <2; (ii) mapping quality >20; (iii) depth of the variant position >3.
Genome-wide association study (GWAS)
All GWAS analyses were performed using Plink (Purcell et al., 2007) software. The SNPS of autosomes and sex chromosomes were analyzed separately using the method of previous studies (O’Sullivan et al., 1991). Since EF/LF trait was a categorical variable, the case/control model was used to analyze the differences in the frequencies of alleles at SNP loci between groups K and M. The Bonferroni method was used to correct for multiple comparisons to obtain statistically significant differences (P < 1.32e-07).
SNPs loci detection
We designed primers to verify the three significant SNPs loci on Z chromosome, aiming to find a reference method for the molecular identification of sex. All primers sets were synthesized by Sangon Biotech Primer Design Center (Shanghai, China) (Supplementary Table SI). First generation sequencing is used for sequence determination.
Results
Genome sequencing data analysis
A total of about 331 Gb of raw data was obtained, and the Quality score of raw reads was concentrate mainly on 36, and most of them >30 (Fig. 2A and Supplementary Table SII). Base composition of reads: base composition deviation occurred in the first 1-10bp; <10bp, the frequencies of the four bases were similar, and there was little difference in position (Fig. 2B). Reads without unique read alignments were discarded; the difference between A/T ratio and G/C ratio at any location was less than 20%.
SNP detection and type distribution
The rare SNPs, loci and SNPs with high sample missing rate in the population will affect the accuracy of subsequent population genetic analysis and GWAS analysis. SNPs were detected, and their quality was filtered using the Vcf tool and Plink software. In total, 11626 high quality SNPs were obtained, which were distributed in 33 autosomes and Z chromosomes. The number of SNPs per chromosome is shown in Supplementary Table SIII. The position of SNP on chromosome is shown in Figure 3, in which exons are 776, accounting for 3.35%, and introns are the most, accounting for 55.32%. Subsequently, 14 major autosomes and Z chromosomes were selected, and the density distribution of SNP in these chromosomes was analyzed in detail (Fig. 4).
Genome-wide association study
We used the Plink software (Version 1.90b3.34; Purcell et al., 2007) for the genome-wide association study. 10 significant SNPs were found in the whole genome, among which the three points with the strongest signal were all on the Z chromosome and located in the genomic regions known to be related to this trait (Fig. 5). For these 10 significant SNP loci, we studied their specific position on the chromosome, allele frequencies among different groups and other information (Supplementary Table SIV).
Filtering of differentially expressed genes and functional enrichment analysis
According to the 10 significant SNP sites selected, all candidate genes within 1M region around the SNP sites were found, and a total of 87 candidate genes were identified, of which 67 were protein-coding RNAs and 20 were long non-coding RNAs (lncRNA) (Supplementary Table SV). Surprisingly, no known candidate genes were found for the three SNPs on the Z chromosome.
All the candidate genes identified in this study may play a role in the phenotype of chicken feather percentage. In order to understand the function and regulatory mechanism of these genes, GO analysis (Fig. 6, Supplementary Table SVI) and KEGG pathway annotation (Table I) were performed. From GO analysis, we can see that these genes are enriched in the following processes: embryonic forelimb morphogenesis; proteasome ubiquitin independent protein catabolic process; peripheral nervous system neuron development, etc. Furthermore, Candidate genes were enriched not only in common pathways such as MAPK signaling pathway, mTOR signaling pathway, and p53 signaling pathway, but also in C-type lectin receptor signaling pathway and Neuroactive ligand-receptor interaction.
Bayesian model for predicting chicken feather pattern
Firstly, each candidate SNP and population frequency information were determined by referring to the sequencing results (Supplementary Table SVII). Then, the probability of being LF under a certain observed genotype was estimated by Bayesian model:
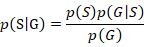
Here, p(G) is the probability of a certain genotype in the whole population, p(S) is the proportion of LF in the population (p (S)=0.5), and p (G│S) is the probability of a certain genotype in the LF. The statistical results of different loci are shown in Table II. Multiple loci can be combined for joint estimation, which provides reference for identification of early and late feather molecules.
Table I. Pathway enrichment analysis of candidate genes.
|
ID |
Gene name |
Kegg_pathway |
|
ALG10 |
ALG10, alpha-1,2-glucosyltransferase (ALG10) |
gga00510: N-Glycan biosynthesis, gga01100: Metabolic pathways, |
|
GTSE1 |
G2 and S-phase expressed 1(GTSE1) |
gga04115: p53 signaling pathway, |
|
PCF11 |
PCF11 cleavage and polyadenylation factor subunit (PCF11) |
gga03015: mRNA surveillance pathway, |
|
AGPS |
alkylglycerone phosphate synthase (AGPS) |
gga00565: Ether lipid metabolism, gga01100: Metabolic pathways, gga04146: Peroxisome, |
|
CALCA |
Calcitonin (CALCA) |
gga04080: Neuroactive ligand-receptor interaction, gga04270: Vascular smooth muscle contraction, |
|
CERK |
ceramide kinase (CERK) |
gga00600: Sphingolipid metabolism, gga01100: Metabolic pathways, |
|
CYP2R1 |
cytochrome P450 family 2 subfamily R member 1(CYP2R1) |
gga00100: Steroid biosynthesis, gga01100: Metabolic pathways, |
|
FZD4 |
frizzled class receptor 4(FZD4) |
gga04150: mTOR signaling pathway, gga04310: Wnt signaling pathway, gga04916: Melanogenesis, |
|
HNRNPA3 |
heterogeneous nuclear ribonucleoprotein A3(HNRNPA3) |
gga03040: Spliceosome, |
|
ME3 |
malic enzyme 3(ME3) |
gga00620: Pyruvate metabolism, gga01100: Metabolic pathways, gga01200: Carbon metabolism, gga03320: PPAR signaling pathway, |
|
NFE2L2 |
nuclear factor, erythroid 2 like 2(NFE2L2) |
gga04141: Protein processing in endoplasmic reticulum, |
|
PDE3B |
phosphodiesterase 3B(PDE3B) |
gga00230: Purine metabolism, gga01100: Metabolic pathways, gga04371: Apelin signaling pathway, gga04910: Insulin signaling pathway, gga04914: Progesterone-mediated oocyte maturation, |
|
PSMA1 |
proteasome subunit alpha 1(PSMA1) |
gga03050: Proteasome, |
|
RRAS |
related RAS viral (r-ras) oncogene homolog(RRAS) |
gga04010: MAPK signaling pathway, gga04137: Mitophagy - animal, gga04140: Autophagy - animal, gga04218: Cellular senescence, gga04371: Apelin signaling pathway, gga04625: C-type lectin receptor signaling pathway, gga04810: Regulation of actin cytoskeleton |
Table II. The probability of EF and LF in different genotypes at each locus.
|
SNP loci |
Gene type |
Fast (%) |
Slow (%) |
|
chrZ: 3583G>C |
GG |
66.12 |
33.88 |
|
GC |
25.81 |
74.19 |
|
|
CC |
1 |
0 |
|
|
chrZ: 6553A>G |
GG |
72.54 |
27.46 |
|
AG |
19.58 |
80.42 |
|
|
AA |
0 |
0 |
|
|
chrZ: 3583A>G |
GG |
72.13 |
27.87 |
|
AG |
12.62 |
87.38 |
|
|
AA |
26.36 |
73.64 |
Discussion
As an important genetic trait in modern poultry breeding, EF/LF phenotype is widely used by poultry breeders all over the world. Not only is it commonly used for sex determination, but it is also associated with growth rate, egg production and disease resistance (O’Sullivan et al., 1991). In addition, the integrity and maturity of feathers is an important factor affecting the carcass beauty of local chickens, especially broilers. At present, it is generally believed that this trait is a pair genetic quality trait, which is controlled by four complex alleles Kn, Ks, K and k on Z chromosome (Dunnington et al., 1986). The late feather (K) is 1dominant to the early feather (k), and the recessive sequence of genes is Kn >Ks >K >k (Somes, 1969). The duplicated gene dPRLR-SPEF2 is localized in the short arm of Z chromosome, which can be used as the molecular basis of EF phenotype (Elferink et al., 2008). Studies have shown that there is no close linkage between it and ev21 endogenous virus insertion (Guo et al., 2014). Therefore, the interlacing situation and degree between late feathering line and early feathering line can be used as the discrimination standard to allow auto-sexing of chicks.
PRLR is involved in the regulation of the development, metabolism and circulation of skin tissues and their appendages (mammary gland, sebaceous gland, hair, etc.) (Craven et al., 2001); and its regulatory role in the growth cycle of animal coat and seasonal hair moult has been widely studied. Similarly, SPEF2 has recently attracted more and more attention as a candidate gene for EF/LF phenotype due to its partial sequence duplication with PRLR (Elferink et al., 2008). However, feather growth is not only associated with the Z chromosome alone, but also affected by related genes on autosomes. In 1946, Jones identified a gene on an autochromosome that controls wing feather growth in chicks and named it tarded, denoted by t. While, many other genes on autosomes that regulate feather growth rate need to be identified and explored. By comparing the genomes of late-feathered chickens and early-feathered chickens, we identified 11626 high-quality SNPs distributed in 33 autosomes and Z chromosomes, which provides a new complement for the study of candidate SNPs related to chicken feather growth.
In this study, we used the Case/Control model to analyze whether there was a significant difference in gene frequencies between the EF and LF phenotype groups at SNP loci. Ten significant SNP loci were found in the whole genome, among which the three points with the strongest signal were all on the Z chromosome and located in the genomic regions known to be related to this trait. The chicken genetic linkage map shows that the K locus is located on the short arm of the Z chromosome and thus can be used for automatic sex identification at hatching, even in the embryonic stage (Hamoen et al., 2001). However, a search of the 1M region around the three SNPS on the Z chromosome yielded no known candidate genes. This may be due to differences in sequencing depth or filtering methods. Interestingly, we found more emerging candidate genes on autosomes that may influence feather development. Among them, Davoodi et al. (2022) identified TBC1D22A located on chromosome 1 as a candidate gene for feather color through 60 k high-density SNP array analysis (Davoodi et al., 2022). Feather color is not only considered a social signal for chickens, but also a breeding recognition tool for breeders. In addition, it has been shown that PRSS23, as a key candidate gene for two different feather follicles, may be involved in the regulation of feather morphology (Xu et al., 2022). Miraculously, among our candidate genes, HOXD family genes were clustered in large numbers, almost including HOXD3- HOXD13. Vertebrate Hox genes encode transcription factors that function in the development of a variety of organs and structures (Schep et al., 2016). Primarily, the HOX gene regulates the development of skin derivatives, such as mammary glands in mammals (Awgulewitsch, 2003; Chuong et al., 1990) and hair follicles and feathers in poultry (Carroll and Capecchi, 2015). Early studies have suggested that the expression of CHOXC-8 and CHOXD-13 in chick skin and skin appendage specifications may be involved in dorsal feather and foot scale formation in chick embryos (Kanzler et al., 1997). Alasdair I Reid found that Hoxb-4, Hoxa-7 and Hoxc-8 were expressed temporally and spatially in a collinear manner in the skin (Reid and Gaunt, 2002). Thus, the HOX gene provides regionally restricted location cues during embryonic skin modeling and also conveys general signals required for feather morphology, which may have a significant impact on transcriptional activity in the developing chick skin.
In this study, we detected multiple pathways that have been confirmed to be related to feather or hair production and development, such as Wnt signaling pathway (Xie et al., 2020), mTOR signaling pathway (Gao et al., 2019), p53 signaling pathway (Qiu et al., 2022), and MAPK signalling pathway (Fang et al., 2018). These pathways not only affect feather development, but also regulate embryonic development, epithelial appendage formation, and hair follicle repair and regeneration. Additionally, we also identified some pathways that have not yet been identified but may also be involved in avian feather development. Metabolic pathways were determined to be associated with the anaerobic degradation of feather keratin and coloration of chicken feathers; Sphingolipid metabolism also acts on quantitative changes in feather color (Wu et al., 2022; Davoodi et al., 2022). Overall, we identified a number of genes and signaling pathways that are associated with or potentially play a role in feather pattern in poultry.
Molecular marker-assisted selection (MAS) is mainly used for breeding new varieties. However, the early identification of sex is equally important for accelerating the speed of breed improvement and commercialization. Bayesian prediction allow to estimate the effect of molecular markers through differential contraction, which can describe the relationship between complex molecular markers and target traits more flexibly (Gianola et al., 2009). Studies have used Bayesian models to identify the relationship between heart-type fatty acid binding proteins and fatty acid metabolism in chickens (Li et al., 2010). In addition, Bayesian models have practical applications in poultry epigenetics and stress (Videla Rodriguez et al., 2022). Here we sequenced three significant SNPs on the Z chromosome using next-generation sequencing; identified candidate SNPs and population frequency information. Roughly, an individual with GC genotype at chrZ: 11263583 locus was found in the population, with 74.19% certainty that the chicken was the LF; 100% of CC genotype was EF trait.
Conclusion
In conclusion, a total of 11626 high-quality SNPs were identified. On this basis, GWAS analysis was carried out to identify molecular genetic markers significantly related to this trait, screen out candidate genes that may be related to feather type, and search for biological pathways to play a role. Therefore, our results may provide a genomic and molecular evidence that autosomes, in addition to sex chromosomes, also have an impact on emergence rate. The combination of population gene frequency information and Bayesian probability model with apparent traits is beneficial to improve the accuracy of early poultry sex discrimination.
Acknowledgments
The authors are grateful to Sichuan Animal Science Academy and Farm Animal Genetic Resources Exploration and Innovation Key Laboratory of Sichuan Province for its availability and support.
Funding
This study was supported by the National Modern Agricultural Technology System Construction of China (CARS-41), Sichuan Science and Technology Program (2021YFYZ0031, 2021YFYZ0009), Sichuan Province Science and Technology Support Program (Grant No. 2021YFYZ0031 and 2021ZHFP0163), National Key Research and Development Project (2022YFD1601608), Sichuan Province Science and Technology Innovation Talent Project (2023JDRC0045).
IRB approval
All animal experiments were approved by the Institutional Animal Care and Use Committee of Sichuan Agricultural University (Certification No. SYXK2019-187).
Ethical statement
The animals and experimental methods used in the experiments were studied in accordance with the guidelines of the Ministry of Agriculture of China. The procedures followed in animal experiments in this study were approved by the Animal Care and Use Committee of Sichuan Academy of Animal Sciences. Strive to minimize the suffering of animals.
All animal experiments were approved by the Institutional Animal Care and Use Committee of Sichuan Agricultural University (Certification No. SYXK2019-187).
There is supplementary material associated with this article. Access the material online at: https://dx.doi.org/10.17582/journal.pjz/20230302070341
Statement of conflict of interest
The authors have declared no conflict of interest.
References
Awgulewitsch, A., 2003. Hox in hair growth and development. Naturwissenschaften, 90: 193-211. https://doi.org/10.1007/s00114-003-0417-4
Bu, G., Huang, G., Fu, H., Li, J., Huang, S. and Wang, Y., 2013. Characterization of the novel duplicated PRLR gene at the late-feathering K locus in Lohmann chickens. J. Mol. Endocrinol., 51: 261-276. https://doi.org/10.1530/JME-13-0068
Carroll, L.S. and Capecchi, M.R., 2015. Hoxc8 initiates an ectopic mammary program by regulating Fgf10 and Tbx3 expression and Wnt/β-catenin signaling. Development, 142: 4056-4067. https://doi.org/10.1242/dev.128298
Chen, S., Zhou, Y., Chen, Y. and Gu, J., 2018. Fastp: An ultrafast all-in-one FASTQ preprocessor. Bioinformatics, 34: i884-i890. https://doi.org/10.1093/bioinformatics/bty560
Chuong, C.M., Oliver, G., Ting, S.A., Jegalian, B.G., Chen, H.M. and De Robertis, E.M., 1990. Gradients of homeoproteins in developing feather buds. Development, 110: 1021-1030. https://doi.org/10.1242/dev.110.4.1021
Collins, W.M., Teeri, A.E., Kahn, A.C., Zervas, N.P. and Costantino, R.F., 1970. The influence of sex-linked barring and rate of feathering genes, and of stock, upon egg riboflavin and vitamin B12. Poult. Sci., 49: 432-439. https://doi.org/10.3382/ps.0490432
Craven, A.J., Ormandy, C.J., Robertson, F.G., Wilkins, R.J., Kelly, P.A., Nixon, A.J. and Pearson, A.J., 2001. Prolactin signaling influences the timing mechanism of the hair follicle: Analysis of hair growth cycles in prolactin receptor knockout mice. Endocrinology, 142: 2533-2539. https://doi.org/10.1210/endo.142.6.8179
Danecek, P., Auton, A., Abecasis, G., Albers, C.A., Banks, E., Depristo, M.A., Handsaker, R.E., Lunter, G., Marth, G.T., Sherry, S.T., Mcvean, G. and Durbin, R., 2011. The variant call format and VCFtools. Bioinformatics, 27: 2156-2158. https://doi.org/10.1093/bioinformatics/btr330
Davoodi, P., Ehsani, A., Vaez Torshizi, R. and Masoudi, A.A., 2022. New insights into genetics underlying of plumage color. Anim. Genet., 53: 80-93. https://doi.org/10.1111/age.13156
Dunnington, E.A., Siegel, P.B. and Gross, W.B., 1986. Sex-linked feathering alleles (K, k+) in chickens of diverse genetic backgrounds. Avian Pathol., 15: 139-148. https://doi.org/10.1080/03079458608436273
Elferink, M.G., Vallée, A.A., Jungerius, A.P., Crooijmans, R.P. and Groenen, M.A., 2008. Partial duplication of the PRLR and SPEF2 genes at the late feathering locus in chicken. BMC Genomics, 9: 391. https://doi.org/10.1186/1471-2164-9-391
Fang, G., Jia, X., Li, H., Tan, S., Nie, Q., Yu, H. and Yang, Y., 2018. Characterization of microRNA and mRNA expression profiles in skin tissue between early-feathering and late-feathering chickens. BMC Genomics, 19: 399. https://doi.org/10.1186/s12864-018-4773-z
Fotsa, J.C., Mérat, P. and Bordas, A., 2001. Effect of the slow (K) or rapid (k+) feathering gene on body and feather growth and fatness according to ambient temperature in a Leghorn x brown egg type cross. Genet. Sel. Evol., 33: 659-670. https://doi.org/10.1186/1297-9686-33-6-659
Fuentes-Pardo, A.P. and Ruzzante, D.E., 2017. Whole-genome sequencing approaches for conservation biology: Advantages, limitations and practical recommendations. Mol. Ecol., 26: 5369-5406. https://doi.org/10.1111/mec.14264
Gao, Q., Zhou, G., Lin, S. J., Paus, R. and Yue, Z., 2019. How chemotherapy and radiotherapy damage the tissue: Comparative biology lessons from feather and hair models. Exp. Dermatol., 28: 413-418. https://doi.org/10.1111/exd.13846
Gianola, D., De Los Campos, G., Hill, W.G., Manfredi, E. and Fernando, R., 2009. Additive genetic variability and the Bayesian alphabet. Genetics, 183: 347-363. https://doi.org/10.1534/genetics.109.103952
Goger, H., Demirtas, S.E., and Yurtogulla, S., 2017. Determination effects of slow (K) and fast (k+) feathering gene on egg production and hatching traits in laying hens. Asian J. Anim. Vet. Adv., 12: 247–253. https://doi.org/10.3923/ajava.2017.247.253
Groenen, M.A., Megens, H.J., Zare, Y., Warren, W.C., Hillier, L.W., Crooijmans, R.P., Vereijken, A., Okimoto, R., Muir, W.M. and Cheng, H.H., 2011. The development and characterization of a 60K SNP chip for chicken. BMC Genomics, 12: 274. https://doi.org/10.1186/1471-2164-12-274
Guo, F., Yang, B., Ju, Z.H., Wang, X.G., Qi, C., Zhang, Y., Wang, C.F., Liu, H.D., Feng, M.Y., Chen, Y., Xu, Y.X., Zhong, J.F. and Huang, J.M., 2014. Alternative splicing, promoter methylation, and functional SNPs of sperm flagella 2 gene in testis and mature spermatozoa of Holstein bulls. Reproduction, 147: 241-252. https://doi.org/10.1530/REP-13-0343
Hamoen, F.F., Van Kaam, J.B., Groenen, M.A., Vereijken, A.L. and Bovenhuis, H., 2001. Detection of genes on the Z-chromosome affecting growth and feathering in broilers. Poult. Sci., 80: 527-534. https://doi.org/10.1093/ps/80.5.527
Kanzler, B., Prin, F., Thelu, J. and Dhouailly, D., 1997. CHOXC-8 and CHOXD-13 expression in embryonic chick skin and cutaneous appendage specification. Dev. Dyn., 210: 274-287. https://doi.org/10.1002/(SICI)1097-0177(199711)210:3<274::AID-AJA8>3.0.CO;2-D
Li, H. and Durbin, R., 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics, 25: 1754-1760. https://doi.org/10.1093/bioinformatics/btp324
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., Marth, G., Abecasis, G. and Durbin, R., 2009. The sequence alignment/map format and SAM tools. Bioinformatics, 25: 2078-2079. https://doi.org/10.1093/bioinformatics/btp352
Li, H., Wu, G., Zhang, J. and Yang, N., 2010. Identification of the heart-type fatty acid-binding protein as a major gene for chicken fatty acid metabolism by Bayesian network analysis. Poult. Sci., 89: 1825-1833. https://doi.org/10.3382/ps.2010-00699
Luo, C., Shen, X., Rao, Y., Xu, H., Tang, J., Sun, L., Nie, Q. and Zhang, X., 2012. Differences of Z chromosome and genomic expression between early and late-feathering chickens. Mol. Biol. Rep., 39: 6283-6288. https://doi.org/10.1007/s11033-012-1449-7
Merkley, J.W. and Lowe, P.C., 1988. Association of rate of feathering genotypes in broilers with production and carcass composition traits. 2. Effect of genotypes and diet on processing traits and lipid deposition. Poult. Sci., 67: 914-919. https://doi.org/10.3382/ps.0670914
Onteru, S.K., Gorbach, D.M., Young, J.M., Garrick, D.J., Dekkers, J.C. and Rothschild, M.F., 2013. Whole genome association studies of residual feed intake and related traits in the pig. PLoS One, 8: e61756. https://doi.org/10.1371/journal.pone.0061756
O’sullivan, N.P., Dunnington, E.A., Smith, E.J., Gross, W.B. and Siegel, P.B., 1991. Performance of early and late feathering broiler breeder females with different feeding regimens. Br. Poult. Sci., 32: 981-995. https://doi.org/10.1080/00071669108417423
Pal, S.K. and Singh, H., 1997. Inheritance of wing feather development rate in guinea fowl (Numida meleagris). Br. Poult. Sci., 38: 245-248. https://doi.org/10.1080/00071669708417980
Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M.A., Bender, D., Maller, J., Sklar, P., De Bakker, P.I., Daly, M.J. and Sham, P.C., 2007. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet., 81: 559-575. https://doi.org/10.1086/519795
Qiu, M., Yu, C., Zhu, S., Liu, S., Peng, H., Xiong, X., Chen, J., Jiang, X., Du, H., Li, Q., Zhang, Z. and Yang, C., 2022. RNA sequencing reveals lncRNA-mediated non-mendelian inheritance of feather growth change in chickens. Genes Genomics, 44: 1323-1331. https://doi.org/10.1007/s13258-022-01304-2
Reid, A.I. and Gaunt, S.J., 2002. Colinearity and non-colinearity in the expression of Hox genes in developing chick skin. Int. J. Dev. Biol., 46: 209-215.
Schep, R., Necsulea, A., Rodríguez-Carballo, E., Guerreiro, I., Andrey, G., Nguyen Huynh, T.H., Marcet, V., Zákány, J., Duboule, D. and Beccari, L., 2016. Control of Hoxd gene transcription in the mammary bud by hijacking a preexisting regulatory landscape. Proc. natl. Acad. Sci. U. S. A., 113: E7720-e7729. https://doi.org/10.1073/pnas.1617141113
Sharma, A., Lee, J.S., Dang, C.G., Sudrajad, P., Kim, H.C., Yeon, S.H., Kang, H.S. and Lee, S.H., 2015. Stories and challenges of genome wide association studies in livestock. A review. Asian-Australas. J. Anim. Sci., 28: 1371-1379. https://doi.org/10.5713/ajas.14.0715
Somes, R.G., Jr., 1969. Delayed feathering, a third allele at the K locus of the domestic fowl. J. Hered., 60: 281-286. https://doi.org/10.1093/oxfordjournals.jhered.a107994
Song, S.F., Kang, X.T., Sun, G.R., Wang, Y.B. and Zhu, X.J., 2003. Study on the breeding of pure lines of fast and slow feather of gushi chicken and the effect of self-separation between male and female. J. Huazhong Agric. Coll., 22: 4. (In Chinese).
Videla Rodriguez, E.A., Pértille, F., Guerrero-Bosagna, C., Mitchell, J.B.O., Jensen, P. and Smith, V.A., 2022. Practical application of a Bayesian network approach to poultry epigenetics and stress. BMC Bioinf., 23: 261. https://doi.org/10.1186/s12859-022-04800-0
Visscher, P.M., Brown, M.A., Mccarthy, M.I. and Yang, J., 2012. Five years of GWAS discovery. Am. J. Hum. Genet., 90: 7-24. https://doi.org/10.1016/j.ajhg.2011.11.029
Wang, Y., Ding, X., Tan, Z., Ning, C., Xing, K., Yang, T., Pan, Y., Sun, D. and Wang, C., 2017. Genome-wide association study of piglet uniformity and farrowing interval. Front. Genet., 8: 194. https://doi.org/10.3389/fgene.2017.00194
Wu, W., Ma, S., Chen, R., Huang, Y. and Deng, Y., 2022. Genome-wide analysis of Keratinibaculum paraultunense strain KD-1(T) and its key genes and metabolic pathways involved in the anaerobic degradation of feather keratin. Arch. Microbiol., 204: 634. https://doi.org/10.1007/s00203-022-03226-9
Xie, H., Zhou, T., Liu, F., Zhou, Y., Qiu, X., Song, Q. and Shi, R., 1985. Studies on breeding and feathering type of the pure line of the early and late feathering of Chengdu white chicken. J. Sichuan Agric. Coll., 3: 9–14. (In Chinese).
Xie, W.Y., Chen, M.J., Jiang, S.G., Yan, H.C., Wang, X.Q. and Gao, C.Q., 2020. The Wnt/β-catenin signaling pathway is involved in regulating feather growth of embryonic chicks. Poult. Sci., 99: 2315-2323. https://doi.org/10.1016/j.psj.2020.01.002
Xu, Q., Xi, Y., Ma, S., Wang, J., Li, J., Han, C., Li, L., Wang, J. and Liu, H., 2022. Transcriptome profiling of morphogenetic differences between contour and flight feathers in duck. Br. Poult. Sci., 63: 597-604. https://doi.org/10.1080/00071668.2022.2026292
Zhao, J., Yao, J., Li, F., Yang, Z., Sun, Z., Qu, L., Wang, K., Su, Y., Zhang, A., Montgomery, S.A., Geng, T. and Cui, H., 2016. Identification of candidate genes for chicken early- and late-feathering. Poult. Sci., 95: 1498-1503. https://doi.org/10.3382/ps/pew131
To share on other social networks, click on any share button. What are these?










