Genome Characterization of Probiotic Lactobacillus delbrueckii Subsp. bulgaricus Strain NDO2 Isolated from Traditional Yogurt using High Throughput Next Generation Sequencing
Genome Characterization of Probiotic Lactobacillus delbrueckii Subsp. bulgaricus Strain NDO2 Isolated from Traditional Yogurt using High Throughput Next Generation Sequencing
Mati Ullah1*, Muhammad Rizwan2, Ali Raza3, Xianlin Zhao4, Yanling Sun4, Sarah Gul5, Muhammad Ihtesham Waheed6, Muhammad Nadeem Khan7, Alvina Gul8, Sami Ullah Jan9* and Chao Huang4*
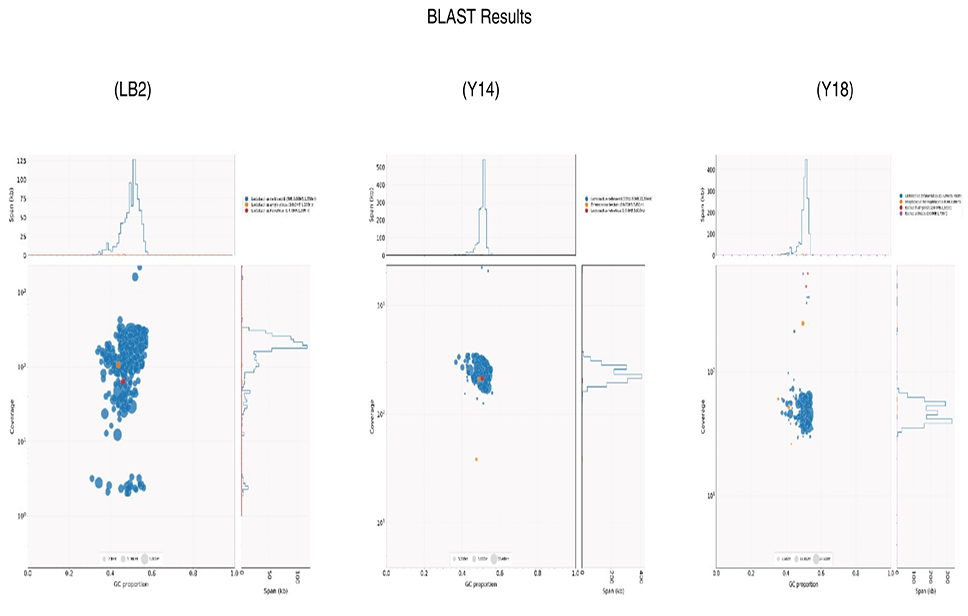
Blob plots indicate the results of BLAST matches of each of the three assembled genomes. The colors indicate BLAST matches to different species of bacteria. The proportion of GC contents in each assembled genome is also indicated.
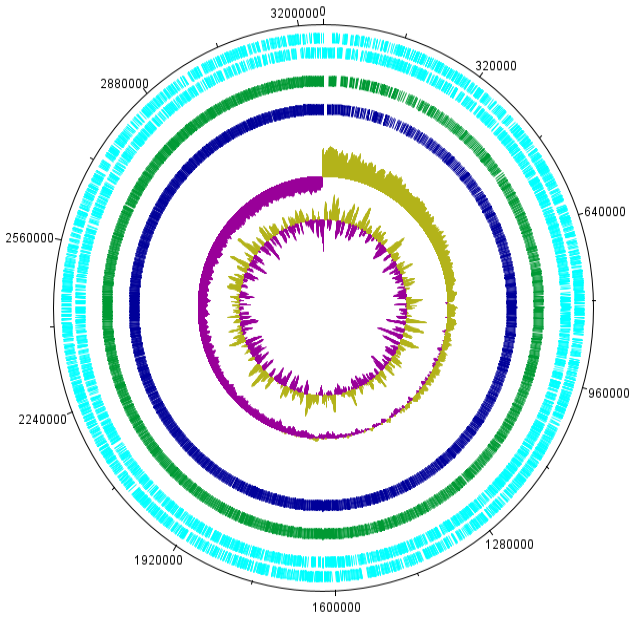
Circular plot of Lactobacillus delbrueckii subsp. Bulgaricus strain NDO2 genome generated by DNA plotter. Circles indicate, from inside outwards: GC skew; GC content; Y18 Comparison results; Y14 comparison result; CDS sequence are (reverse frame); CDS sequence area (plus frame).
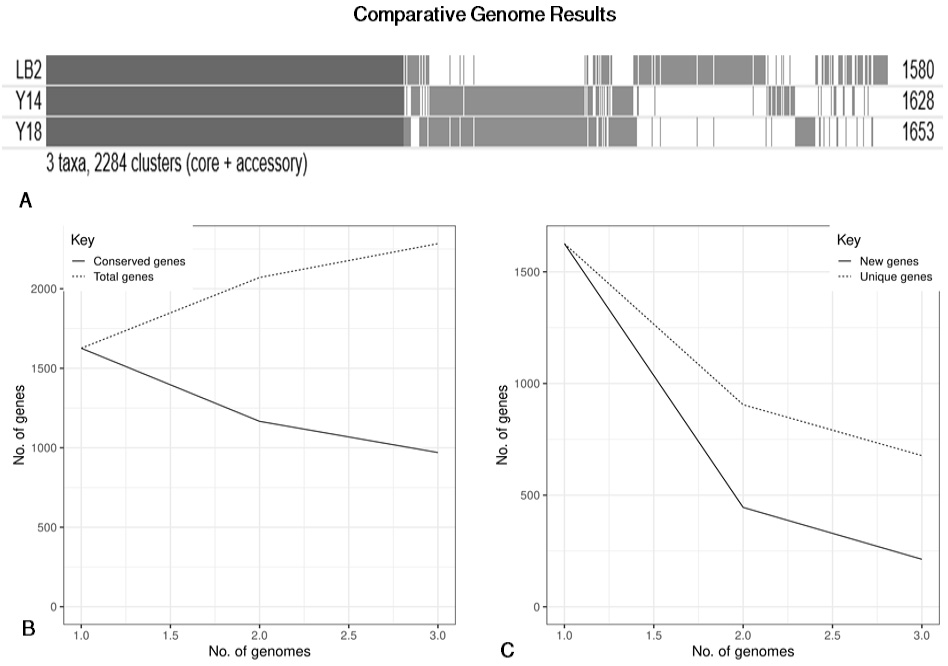
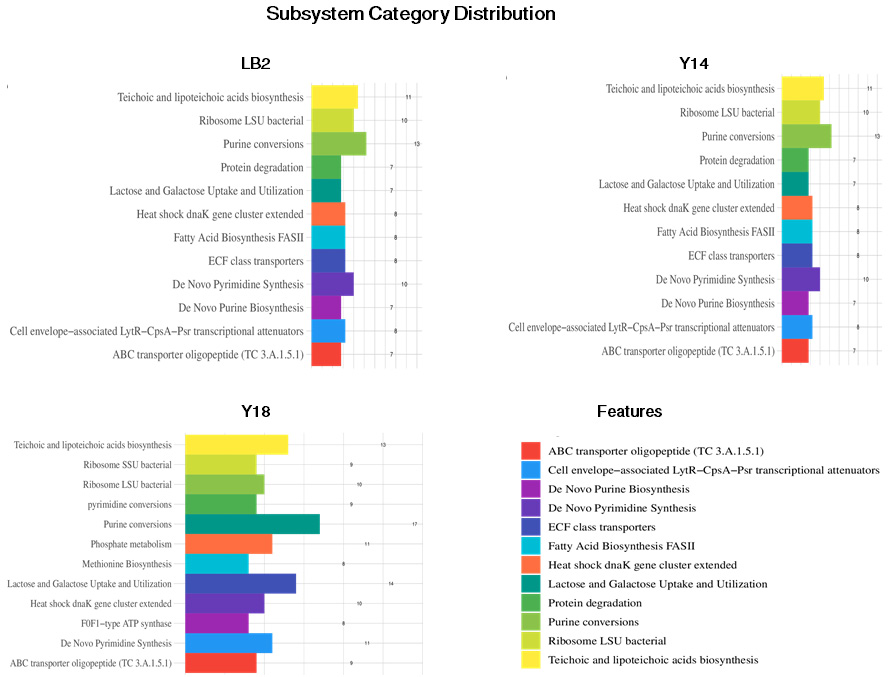
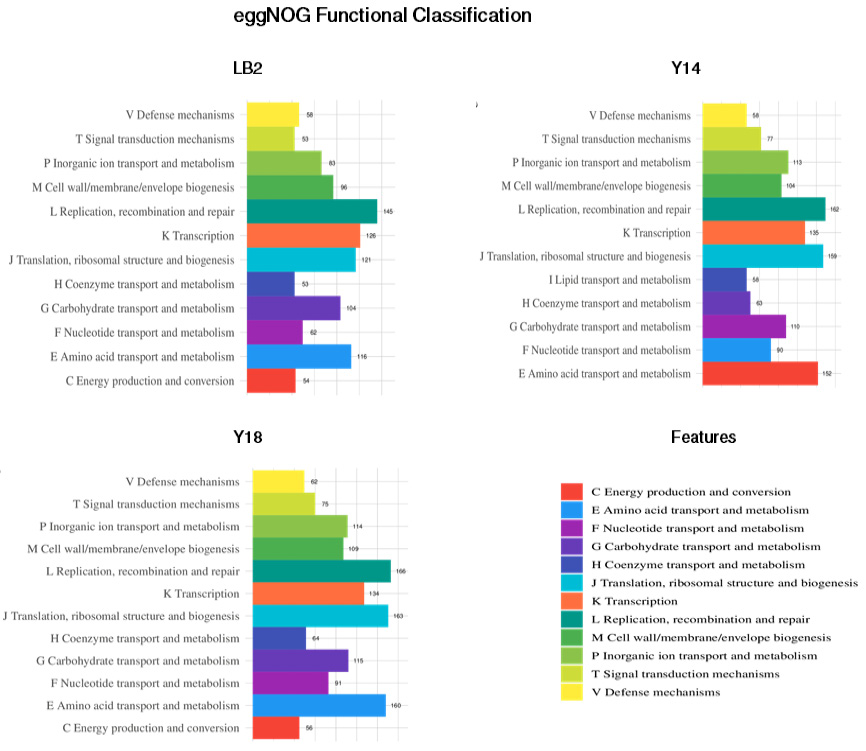
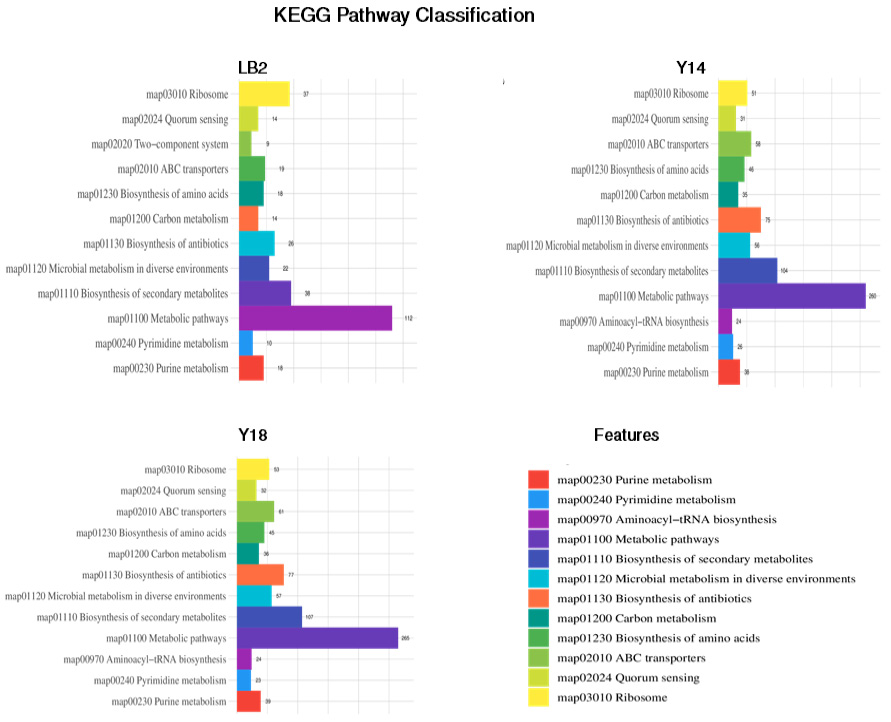
Classification of the KEGG pathways based on the assembled genome of the three probiotic bacterial isolates. The different colors represent the various KEGG pathways identified in the current analysis.