Effects of Florfenicol Stress on 16S rDNA Sequence Diversity of Soil Phosphorus Solubilizing Bacteria
Effects of Florfenicol Stress on 16S rDNA Sequence Diversity of Soil Phosphorus Solubilizing Bacteria
Songruo Tao1, Cuiyi Liao1, Jinju Peng1, Yuexia Ding1,* and Yi Ma1,2,*
1Department of Veterinary Medicine, College of Coastal Agricultural Sciences, Guangdong Ocean University, Zhanjiang 524088, China
2Maoming Branch, Guangdong Laboratory for Lingnan Modern Agriculture, Maoming, 525000, China
ABSTRACT
Florfenicol has a significant therapeutic effect on animal bacterial diseases, most of metabolites enter the soil in the form of metabolites in feces or urine and pollute environment. Soil ecological model was established by experiment to investigate the effects of florfenicol residues in soil on 16S rDNA sequence diversity of phosphorus-solubilizing bacteria. Five different concentrations of florfenicol (0 mg·kg-1, 0.1 mg·kg-1, 1 mg·kg-1, 10 mg·kg-1 and 100 mg·kg-1) were used to collect soil samples on the 7 d, 21 d and 49 d after dosing. The effects of florfenicol on 16S rDNA sequence diversity of soil phosphorus-solubilizing bacteria were determined by amplifed ribosomal DNA restriction analysis (ARDRA) and enterobacterial repetitive intergenic consensus-polymerase chain reaction (ERIC-PCR) method. The results showed that the number of Operational Taxonomic Units (OTUs) types decreased with the increase of florfenicol concentration, and the percentage of OTUs number to bacteria were the lowest at 100 mg·kg-1 florfenicol concentration after 21d treating, which was 8.33%. The phosphorus-solubilizing bacteria were amplified by ERIC-PCR after 21d treating, the fingerprint type of ERIC-PCR decreased with the increase of drug concentration, and the diversity index of the drug group was significantly lower than that of the blank control group. This indicated that florfenicol had an effect on the dominance, richness and evenness of soil phosphorus-solubilizing bacteria community.
Article Information
Received 13 November 2021
Revised 23 December 2021
Accepted 05 January 2022
Available online 12 May 2022
(early access)
Published 27 February 2023
Authors’ Contribution
ST and YD presented the concept of the study. CL and ST curated the data.
Songruo Tao: Methodology; Writing-original draft. JP and CL did formal analysis. CL planned the methodology. ST, JP, YD and YM wrote the manuscript. YD and YM supervised the project. YM managed funds acquisition.
Key words
Florfenicol, Residue, Phosphorus-solubilizing bacteria, 16S rDNA, Diversity
DOI: https://dx.doi.org/10.17582/journal.pjz/20211113031104
* Corresponding author: dingyuexia2006@163.com, mayi761@163.com
0030-9923/2023/0003-1147 $ 9.00/0
Copyright 2023 by the authors. Licensee Zoological Society of Pakistan.
This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
INTRODUCTION
The soil microbial species are large and complex extremely. Phosphorus-solubilizing bacteria as one of the functional microbial groups, which can secrete a variety of organic acids and enzymes to convert insoluble phosphates into usable form (Alori et al., 2017), thus promoting the growth and development of plants and being an important member of the soil phosphorus cycle (Kannan et al., 2021). The distribution and quantity of phosphorus-solubilizing bacteria are related to plant species, soil environment and human disturbance. The microorganisms with phosphorus-solubilizing ability included bacteria, fungi and actinomycetes, and more than 20 genera of phosphorus-solubilizing bacteria had been Areported, and the species and quantity of bacteria accounted for the majority among them (Sharma et al., 2013). In recent years, the diversity of soil microbial community structure had been destroyed seriously by the extensive use of antibiotics (Kotzerke et al., 2011; Liao et al., 2019). t present, isolation and identification of bacteria, phosphorus-solubilizing ability and plant growth are focused on the studies about phosphorus- solubilizing bacteria (Parastesh et al., 2019), however, there are no reports on the impacts on the impact of antibiotics on the community structure of phosphorus-solubilizing bacteria.
Florfenicol has a significant therapeutic effect on animal bacterial diseases in livestock, poultry and aquatic animals (Dinos et al., 2016). It is often applied in veterinary clinic as veterinary antibiotic. In 2013, the widespread use of florfenicol in China reached nearly 10,000 tons, ranking second among all veterinary antibiotics (Zhang et al., 2015). However, florfenicol may be causing pollution, because of difficult to be completely digested and absorbed, and most of it will enter the soil environment in the form of metabolites in feces or urine. Zong et al. (2010) believed that florfenicol residue could lead to changes in microbial communities in the environment, florfenicol was detected in 6 of 11 seawater samples affected by aquaculture discharge, with concentrations of 64.2 μg·L-1, 390.6 μg·L-1, 1.1×104 μg·L-1, 29.8 μg·L-1, 61.6 μg·L-1, 34.9 μg·L-1, respectively. It has been reported that florfenicol degrades slowly in soil, such as Xu added 5 mg·kg-1florfenicol to three kinds of soil (soil from nanchang, hangzhou, changchun) after sterilization and sterilization, respectively. The degradation rate of florfenicol in unsterilized soil was close to 100% after 60 days, florfenicol was more stable in neutral soils than in acidic or alkaline soils, with half-lives of 8.1 d, 7.0 d and 8.5 d in three soils (Nanchang, Hangzhou and Changchun), respectively (Xu et al., 2015). Currently, florfenicol affected the phosphorus-solubilizing bacteria community structure was unclear. In this experiment, soil ecological model was established and phosphorus-solubilizing bacteria were isolated. ARDRA and ERIC-PCR were used to investigate the effects of florfenicol residues on the diversity of 16S rDNA sequence of phosphorus-solubilizing bacteria, in order to provide a theoretical basis for the ecological risks caused by antimicrobial residues in the environment.
MATERIALS AND METHODS
The selected soil
The source of the soil was high-quality brown and yellow soil with a depth of about 20 cm in the campus of Guangdong Ocean University, fertile and suitable for growing vegetables and flowers and other plants, no veterinary drug residue. The physical and chemical properties of soil were as follows: PH 5.42, Alkali-hydrolyzed nitrogen 22.16 mg·kg-1, Salinity 88 us·cm-1, Total nitrogen 0.55 mg·kg-1, Available potassium 263.25 mg·kg-1, Available phosphorus 46.80 mg·kg-1, Organic matter 10.97 mg·kg-1.
Drug and reagents
Florfenicol (Content of 98%) was provided by Shandong Guobang Pharmaceutical Co., LTD. (Batch number: 701-2005101). Inorganic phosphorus liquid culture medium and solid culture medium (PVK) were purchased from Qingdao Haibo Biotechnology Co., LTD. LB medium was purchased from Beijing Luqiao Technology Co., LTD. Premix LA Taq™ Version 2.0, DNA Marker, restriction enzyme Msp I and Afa I were purchased from Takara. The primers were designed and provided by Sangon Bioengineering (Shanghai) Co., LTD.
Soil treatment and sample collection
The collected fresh soil was dried and filtered through a 4 mm-sieve in the shade. The soil divided into 5 groups, 3 replications were performed for each treatment. The soil samples were treated with florfenicol to 0 mg·kg-1, 0.1 mg·kg-1, 1 mg·kg-1, 10 mg·kg-1 and 100 mg·kg-1, respectively. Soil moisture content was adjusted to 50%, which was the maximum water holding capacity in the field and cultured at 26℃. Soil samples were collected from each group on day 7, 21 and 49 after dosing, and bacterial suspension was prepared by 10-fold dilution method.
Bacterial suspension was coated on PVK solid medium, and cultured at 32℃ for 3 days. Single colonies with transparent phosphorus-soluble cycles were selected and inoculated in LB liquid medium at 32℃ and 180 rpm for overnight, then stored at -20℃ with 30% glycerol.
DNA template preparation
The phosphorus-solubilizing bacteria were inoculated into 2 mL of LB liquid medium, cultured at 30℃ for 18 h with shaking at 180 rpm. Following centrifugation at 12000 rpm for 5 min, the supernatant was discard and 1 mL sterile water was added, centrifuged at 12000 rpm for 5 min after mixing fully. Finally, 100 μL deionized water was added and boiled at 100℃ for 10 min, placed on ice for 3 min, centrifuged at 12000 rpm for 5 min, supernatant was absorbed and stored at -20℃ in a new centrifugal tube.
PCR amplification of 16S rDNA
The following universal primers were used.
27 F: 5’-AGAGTTTGATCCTGGCTCAG-3’ and
1492 R: 5’-ACGGTTACCTTGTTACGACTT-3’.
A total of 15 μL reaction including Taq polymerase 13 μL, primers 0.5 μL for each and DNA 1 μL was set up with the reaction parameters of 94℃ 5 min, 30 cycles of 94℃ 30 sec, 56℃ 30 sec, and 72℃ 45 sec, followed by extension for 10 min. PCR product was visualized after electrophoresis.
Digestion of the PCR product
Restriction enzymes Msp I and Afa I were used for the digestion of the PCR product with the following reaction. PCR product 8 μL, Msp I 1 μL, Afa I 1 μL, 10×buffer 2 μL, 0.1%BSA 2 μL and deionized water 6 μL. After incubation at 37℃ for 3 h, 10×loading buffer was added to terminate the reaction, and electrophoresis at 1.8% agarose was performed before imaging.
ERIC-PCR fingerprint analysis
ERIC specific primers were used.
F: 5’-ATGTAAGCTCCTGGGGATTCAC-3’ and
R: 5’-AAGTAAGTGACTGGGGTGAGCG-3’.
A total of 20 μL reaction including Premix LA Taq polymerase 10 μL, primers 1 μL for each, DNA 2 μL and ddH2O 6 μL was set up with the reaction parameters of 94℃ 5 min, 30 cycles of 94℃ 30 sec, 56℃ 30 sec, and 72℃ 45 sec, followed by extension for 10 min. PCR product was visualized after electrophoresis. The amplification bands of each sample were counted.
The community structure diversity of phosphorus-solubilizing bacteria was analyzed by ERIC-PCR fingerprint, and the diversity was measured by the following indicators:
(1) Shannon-Wiener index (H´): ni represents the number of individuals of the i species, N represents the total number of all individuals.
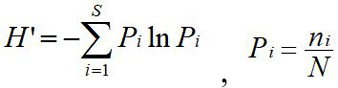
(2) Simpson index (D):
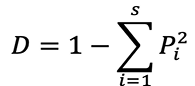
(3) Margalef index (dMa): S represents the number of species in the community.
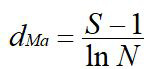
(4) Pielou index(Jsw): Hmax is the maximum species diversity in the community.
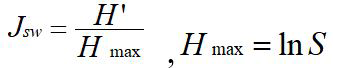
RESULTS
ARDRA analysis
A total of 826 phosphorus-solubilizing bacteria were isolated from soil. PCR amplification of 16S rDNA of phosphorus-soluble bacteria and enzyme digestion of PCR products, ARDRA types can be obtained, and different ARDRA types were derived from different Operational Taxonomic Unit (OTU). Part of the ARDRA atlas was shown in Figure 1. The number of OTUs obtained after enzymatic digestion by phosphorus-solubilizing bacteria was shown in Table I.
The number of OTUs types decreased gradually on day 21 and 49 after dose-adding. When the drug concentration was 100 mg·kg-1, the percentages of OTUs and bacteria were the lowest in the group, which were 8.33% and 9.26%, respectively. After 21 days of treatment, the percentages of OTUs and bacteria were 10.53%, 8.47% and 8.33% in 1, 10 and 100 mg·kg-1 groups, respectively. The sampling time point at which drugs had the greatest influence on phosphorus-solubilizing bacteria ARDRA type was 21d among them.
ERIC-PCR fingerprint analysis
ERIC-PCR was used to amplify phosphorus-solubilizing bacteria on day 21 after dosing, and the fingerprint of bacterial genome structure was obtained. The results of ERIC-PCR amplification and diversity index were shown in Table II. As the concentration of the drug increases, bacteria and types of ERIC-PCR both declined, group I was 65(30), group II was 49(19), group III was 57(18), group IV was 59(14), group V was 60(9), respectively. The Margalef index, Shannon-Wiener index, Simpson index and Pielou index all decreased significantly.
Table I. Analysis of ARDRA patterns of 16S rDNA gene of soil phosphorus solubilizing bacteria.
|
Concentration (mg·kg-1) |
7th d |
21st d |
49th d |
||||||
|
Bacteria |
OTUs |
OTUs/ bacteria |
Bacteria |
OTUs |
OTUs/ bacteria |
Bacteria |
OTUs |
OTUs/ bacteria |
|
|
0 |
61 |
8 |
13.11 |
65 |
8 |
12.31 |
49 |
6 |
12.24 |
|
0.1 |
59 |
8 |
13.56 |
49 |
5 |
10.20 |
50 |
5 |
10.00 |
|
1 |
61 |
7 |
11.48 |
57 |
6 |
10.53 |
56 |
6 |
10.71 |
|
10 |
48 |
5 |
10.42 |
59 |
5 |
8.47 |
50 |
5 |
10.00 |
|
100 |
48 |
6 |
12.50 |
60 |
5 |
8.33 |
54 |
5 |
9.26 |
Table II. Diversity of ERIC-PCR patterns of soil phosphate-solubilizing bacteria after dosing for 21d.
|
Concentration (mg·kg-1) |
Bacteria |
Types of ERIC-PCR |
Margalef index (dMa) |
Shannon-wiener index (H´) |
Simpson index (D) |
Pielou index (Jsw) |
|
0 |
65 |
30 |
6.95 |
3.12 |
0.96 |
0.92 |
|
0.1 |
49 |
19 |
4.63 |
2.59 |
0.82 |
0.88 |
|
1 |
57 |
18 |
4.20 |
2.11 |
0.78 |
0.73 |
|
10 |
59 |
14 |
3.19 |
1.62 |
0.59 |
0.61 |
|
100 |
60 |
9 |
1.95 |
0.98 |
0.39 |
0.45 |
DISCUSSION
ARDRA analysis has been widely used in the study of environmental microbial diversity, which for the analysis of microbial diversity by using restriction endnuclide enzyme digestion of 16S rDNA fragments (Kirk et al., 2004). Recently, Didari et al. (2020) isolated 87 strains halotolerant bacteria from a seasonal high salt lake in Iran, and identified 30 bacterial species by using ARDRA analysis. Wu et al. (2018) isolated 1081 endophytic bacteria from the roots, stems and leaves of Dendrobium nobilis from three sample sites in Guizhou, and identified 41 OTUs using ARDRA analysis method. In this experiment, ARDRA analysis was used to classify 90 OTUs from 826 soil phosphorus- solubilizing bacteria.
Antibiotic residues in the environment would increase the risk of resistance, and the increase in antibiotic concentrations and types may also change the structural diversity and function of microbial communities (Proia et al., 2013). Microbial activity was significantly affected even at very low antibiotic concentrations (Xu et al., 2021). Girardi et al. (2011) explored the degradation of ciprofloxacin in water and soil, and found that ciprofloxacin could significantly inhibit microbial activity and affect microbial community structure, and the inhibition effect in water environment was stronger than that in soil environment. Zou et al. (2018) applied aureomycin mixed with pig manure into soil and found that the relative abundance of dominant bacteria and microbial community structure in soil would be changed. Toth et al. (2011) found that sulfadimethazine could periodically inhibit soil nitrification, and microbial activity would also be significantly affected. In this experiment, different concentrations of florfenicol were added into soil, and it was found that with the increase of drug concentration and the extension of action time, the number of OTUs types of phosphorus-solubilizing bacteria decreased gradually. At the florfenicol concentration of 100 mg·kg-1, the percentage of OTUs and bacteria was the lowest at 8.33% after 21 days of treatment. The results showed that high concentration of florfenicol inhibited the activity of phosphorus-solubilizing bacteria in soil, and thus weakened the diversity of bacteria community.
ERIC-PCR amplification was performed on phosphorus-solubilizing bacteria after 21 days dosing. It was found that the higher the florfenicol concentration, the lower the Margalef index, Shannon-Wiener index, Simpson index and Pielou index, presenting a dose-dependent effect. Fan et al. (2018) explored the effects of colistin sulfate on the genetic diversity of soil denitrifying bacteria nirS and nosZ, and found that the higher the colistin sulfate concentration, the lower the overall diversity of microbial community. Zhang et al. (2014) explored the impact of roxarsone residue on soil microbial community and found that the higher the concentration of roxarsone in soil, the more significant the impact on the structural diversity of soil microbial community.The results showed that under the stress of florfenicol, the community structure diversity of phosphorus-solubilizing bacteria decreased, and the diversity index analysis showed that florfenicol had an effect on the dominance, richness and evenness of phosphorus-solubilizing bacteria community.
Phosphorus-solubilizing bacteria are important microorganisms in phosphorus cycling, which can decompose insoluble phosphate in soil and provide available forms of phosphorus for plant growth and development. However, some studies have found that the phosphorus solubilizing ability of the bacteria become weakened, and some even lose the phosphorous solubilizing ability after rifampicin treatment (Zhang et al., 2015). Veterinary drugs containing heavy metals, such as arsenic preparations, which have an inhibitory effect on soil phosphorus solubilizing bacteria (Van et al., 1976). Florfenicol reduced the community diversity of phosphorus solubilizing bacteria in soil, and affected the dominance, richness and evenness of phosphorus solubilizing bacteria community. Whether it affected the phosphorus solubilizing ability of bacteria needs to be further explored.
CONCLUSION
To summarize, based on the findings of the above experiments, this study obtained 826 phosphorus-solubilizing bacteria from soil. The results of ARDRA analysis showed that high concentration of florfenicol inhibited the activity of phosphorus-solubilizing bacteria, and weakened the diversity of bacteria. ERIC-PCR fingerprint analysis showed that florfenicol stress reduced the diversity of phosphorus-solubilizing bacteria community structure, and diversity index analysis showed that florfenicol had an effect on the dominance, richness and evenness of phosphorus-solubilizing bacteria community.
ACKNOWLEDGMENTS
This work was financially supported by 2019 Guangdong University Features Innovation Project By Department of Education in Guangdong Province, China (2019KTSCX057) and Guangdong Provincial Department of Education 2021Special project for Key fields of Ordinary colleges and Universities (2021ZDZX4003).
Data availability statement
All public data generated or analyzed during this study are included in this article. Data sharing is not applicable to this article as no new data were created or analyzed in this study.
Statement of conflict of interest
The authors have declared no conflict of interest.
REFERENCES
Alori, E.T., Glick, B.R., and Babalola, O.O., 2017. Microbial phosphorus solubilization and its potential for use in sustainable agriculture. Front. Microbiol., 8: 971. https://doi.org/10.3389/fmicb.2017.00971
Didari, M., Bagheri, M., and Amoozegar, M.A., 2020. Diversity of halophilic and halotolerant bacteria in the largest seasonal hypersaline lake (Aran-Bidgol-Iran). Iran. J. environ. Hlth. Sci. Eng., 18: 961-971. https://doi.org/10.1007/s40201-020-00519-3
Dinos, G.P., Athanassopoulos, C.M., Missiri, D.A., Giannopoulou, P.C., Vlachogiannis, I.A., Papadopoulos, G.E., Papaioannou, D., and Kalpaxis, D.L., 2016. Chloramphenicol derivatives as antibacterial and anticancer agents: Historic problems and current solutions. Antibiotics (Basel), 5: 20. https://doi.org/10.3390/antibiotics5020020
Fan, T.L., Sun, Y.X., Peng, J.J., Niu, J.L., Zhong, X.X., Wang, M.Z., and Ma, Y., 2018. Effects of colistin sulphate on nirS,nosZ gene diversity of soil denitrification bacteria. Asian J. Ecotoxicol., 13: 157-165.
Girardi, C., Greve, J., and Lamshöft, M., 2011. Biodegradation of ciprofloxacin in water and soil and its effects on the microbial communities. J. Hazard. Mater., 198: 22-30. https://doi.org/10.1016/j.jhazmat.2011.10.004
Kannan, P., Paramasivan, M., Marimuthu, S., Swaminathan, C., and Jayakumar, B., 2021. Applying both biochar and phosphobacteria enhances Vigna mungo L. growth and yield in acid soils by increasing soil pH, moisture content, microbial growth and P availability. Agric. Ecosyst. Environ., 308: 107258. https://doi.org/10.1016/j.agee.2020.107258
Kirk, J.L., Beaudette, L.A., Hart, M., Moutoglis, P., Klironomos, J.N., Lee, H., and Trevors, J.T., 2004. Methods of studying soil microbial diversity. J. Microbiol. Methods, 58: 169-188. https://doi.org/10.1016/j.mimet.2004.04.006
Kotzerke, A., Hammesfahr, U., Kleineidam, K., Lansnoft, M., Thiele-Bruhn, S., Scholter, M., Wilke, B.M., 2011. Influence of difloxacin-contaminated manure on microbial community structure and function in soils. Biol. Fert. Soils, 47: 177-186. https://doi.org/10.1007/s00374-010-0517-1
Liao, Q., Li, M.Z., and Dong, Y.P., 2019. Impacts of Cu and sulfadiazine on soil potential nitrification and diversity of ammonia-oxidizing archaea and bacteria. Env. Pollut. Bioavail., 31: 60-69. https://doi.org/10.1080/26395940.2018.1564629
Parastesh, F., Alikhani, H.A., and Etesami, H., 2019. Vermicompost enriched with phosphate- solubilizing bacteria provides plant with enough phosphorus in a sequential cropping under calcareous soil conditions. J. Clean. Prod., 221: 27-37. https://doi.org/10.1016/j.jclepro.2019.02.234
Proia, L., Lupini, G., Osorio, V., Perez, S., Barcelo, D., Schwartz, T., Amalfitano, S., Fazi, S., Romani, A.M., and Sabater, S., 2013. Response of biofilm bacterial communities to antibiotic pollutants in a Mediterranean River. Chemosphere, 92: 1126-1135. https://doi.org/10.1016/j.chemosphere.2013.01.063
Sharma, S.B., Sayyed, R.Z., Trivedi, M.H., and Gobi, T.A., 2013. Phosphate solubilizing microbes: sustainable approach for managing phosphorus deficiency in agricultural soils. Springerplus, 2: 587. https://doi.org/10.1186/2193-1801-2-587
Toth, J.D., Feng, Y.C., and Dou, Z.X., 2011. Veterinary antibiotics at environmentally relevant concentrations inhibit soil iron reduction and nitrification. Soil Biol. Biochem., 43: 2470-2472. https://doi.org/10.1016/j.soilbio.2011.09.004
Van, D.P., and van de Voorde, H., 1976. Sensitivity of environmental microorganisms to antimicrobial agents. Appl. environ. Microbiol., 31: 332-336. https://doi.org/10.1128/aem.31.3.332-336.1976
Wu, Q.S., Lei, X., Lei, Y.M., Pu, X., Liu, Y., and Weng, Q.B., 2018. Analyses on composition and diversity of endophytic bacteria in Dendrobium nobile. J. Pl. Resour. Environ., 27: 79-90.
Xu, L.S., Wang, W.Z., and Xu, W.H., 2021. Effects of tetracycline antibiotics in chicken manure on soil microbes and antibiotic resistance genes (ARGs). Environ. Geochem. Hlth., pp. 1-12. https://doi.org/10.1007/s10653-021-01004-y
Xu, M.F., Qian, M.R., Zhang, H., Ma, J.W., Wang, J.M., and Wu, H.Z., 2015. Simultaneous determination of florfenicol with its metabolite based on modified quick, easy, cheap, effective, rugged, and safe sample pretreatment and evaluation of their degradation behavior in agricultural soils. J. Sep. Sci., 38: 211-217. https://doi.org/10.1002/jssc.201400919
Zhang, F., Bai, L., Guo, R.Z., Ma, Y., and Sun, Y.X., 2014. Effects of roxarsone residue on the microbial community structure in soil. Asian J. Ecotoxicol., 9: 475-482.
Zhang, J.J., Li, J.G., Guo, Y.P., Han, C., and Qin, Y.T., 2015. Screening and identifi cation of phosphate-solubilizing bacteria in rhizosphere soil of Xinjiang walnut. Nonwood For. Res., 33: 57-62.
Zhang, Q.Q., Ying, G.G., Pan, C.G., Liu, Y.S., and Zhao, J.L., 2015. Comprehensive evaluation of antibiotics emission and fate in the river basins of China: Source analysis, multimedia modeling, and linkage to bacterial resistance. Environ. Sci. Technol., 49: 6772-6782. https://doi.org/10.1021/acs.est.5b00729
Zong, H.M., Ma, D.Y., and Wang, J.Y., 2010. Research on florfenicol residue in coastal area of Dalian (Northern China) and analysis of functional diversity of the microbial community in marine sediment. Bull. environ. Contamin. Toxicol., 84: 245-249. https://doi.org/10.1007/s00128-009-9923-1
Zou, Y., Huang, W.H., Chen, Y.J., Feng, H.J., Li, J.H., Ren, H.X., and Sun, Y.X., 2018. Microbial community diversity and tetracycline resistance gene abundance in manure-soil model under chlortetracycline stress. J. S. China Agric. Univ., 39: 65-73.
To share on other social networks, click on any share button. What are these?










