Development and Validation of Rapid Multiplex PCR Assay for Identification of DNA Origins of Pork, Donkey and Cow Species in Real Food Samples
Development and Validation of Rapid Multiplex PCR Assay for Identification of DNA Origins of Pork, Donkey and Cow Species in Real Food Samples
Muhammad Safdar1*, Muhammad Younus2, Faiz-ul Hassan1 and Yasmeen Junejo3
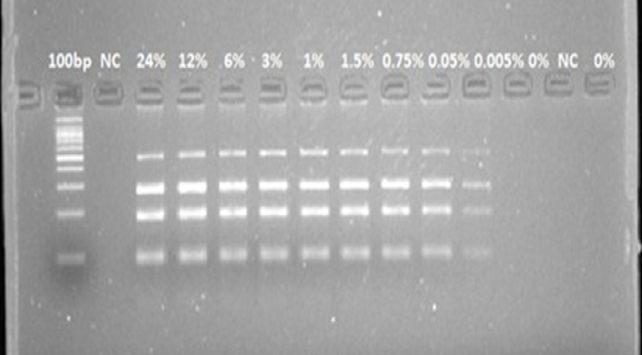
Evaluation of multiplex PCR assay sensitivity for universal, cow, donkey and pork in cooked labortaty prepared samples; M: Marker 100-bp. NC: Negative control (reagents with primers without DNAs); Eu: Eukaryotic DNA positive control. (1) 24%, (2) 12%, (3) 6%, (4) 3%, (5) 1.5%, (6) 0.75%, (7) 0.05% (8) 0.005% (9) 0%.
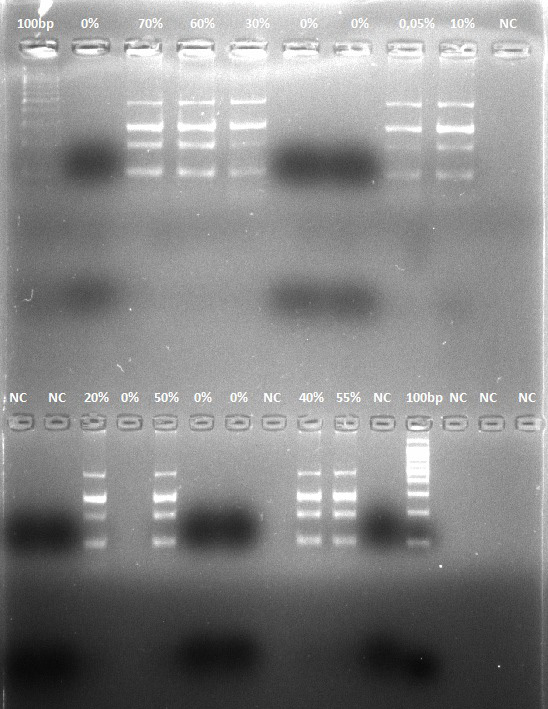
Evaluation of multiplex PCR assay validity was performed on cow, donkey and pork in cooked labortaty prepared samples. M, Marker 100-bp; NC, Negative control (reagents with primers without DNAs).
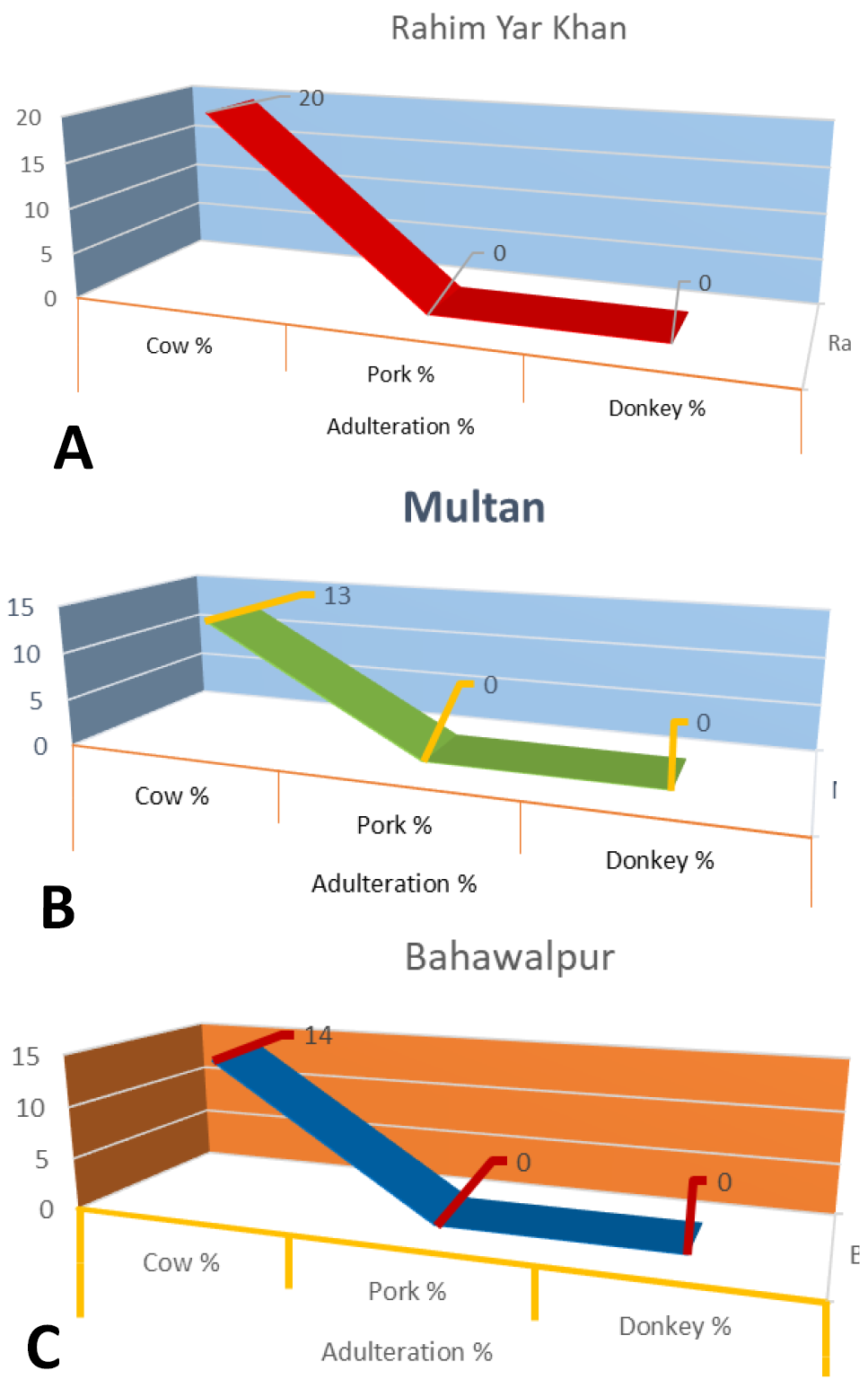
Adulteration in commercial food samples at RYK district (A), Multan district (B) and Bahawalpur district (C).
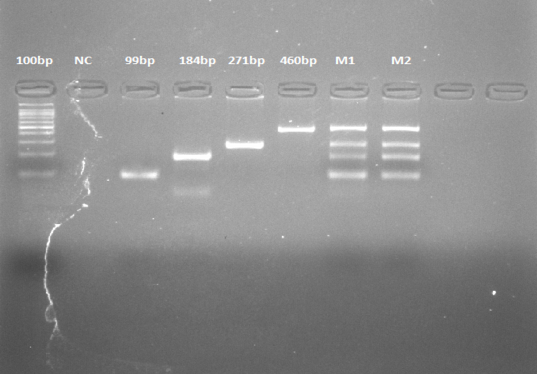
100bp: Ladder, NC: Negative Control (Everything without primer of cow), Cow (Positive Control), Universal primer (99bp), Donkey (184bp), Cow (271bp), Pork (459bp). M1; Multiplex PCR 1, M2; Multiplex PCR 2 (Repeat).