Canine Coronavirus (CCoV), a Neglected Pathogen: Molecular Diversity of S, M, N and 3b Genes
Canine Coronavirus (CCoV), a Neglected Pathogen: Molecular Diversity of S, M, N and 3b Genes
Iracema Nunes de Barros1,2, Luciane Dubina Pinto3, Rosely Bianca dos Santos Kuroda1, Sheila Oliveira de Souza Silva1, Leonardo José Richtzenhain1,2, Cláudio Wageck Canal3 and Paulo Eduardo Brandão1,2*
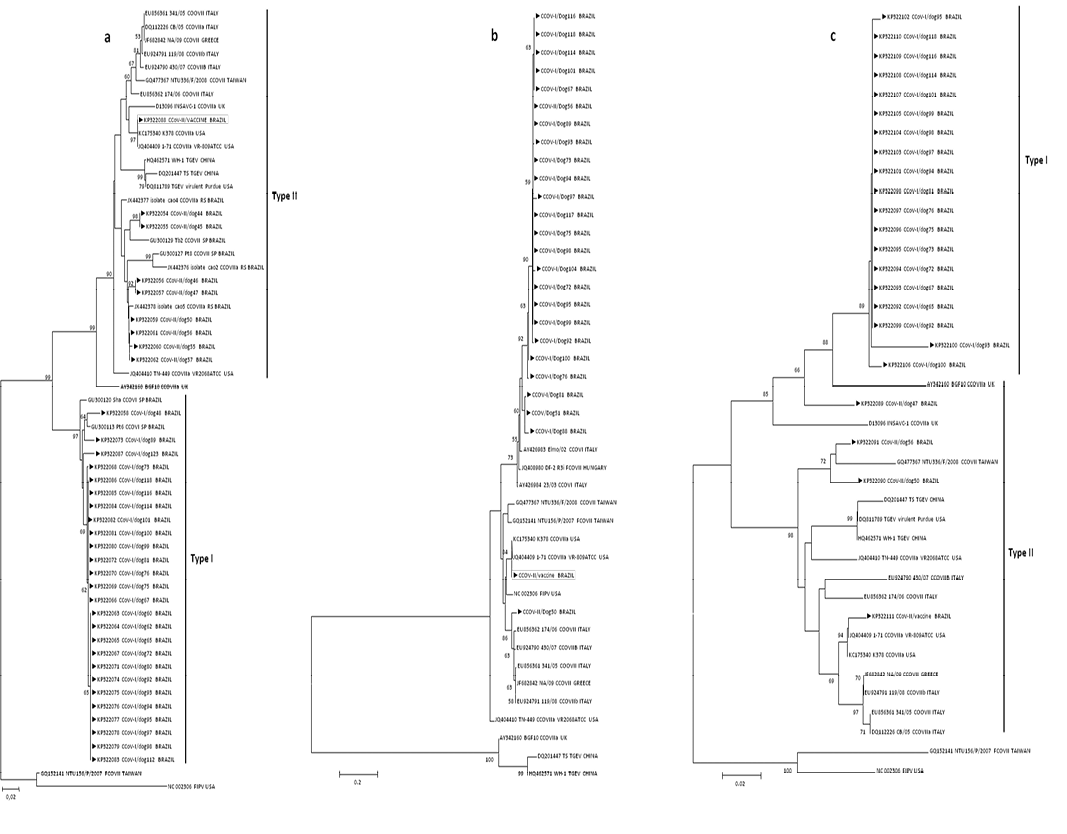
Neighbor-joining nucleotides trees (MCL model) for partial sequences of CCoV genes a: M, b: 3b and c: N; arrows indicate strains form this study. Numbers at each node are bootstrap values (1,000 replicates). The bar represents the number of substitutions per site.
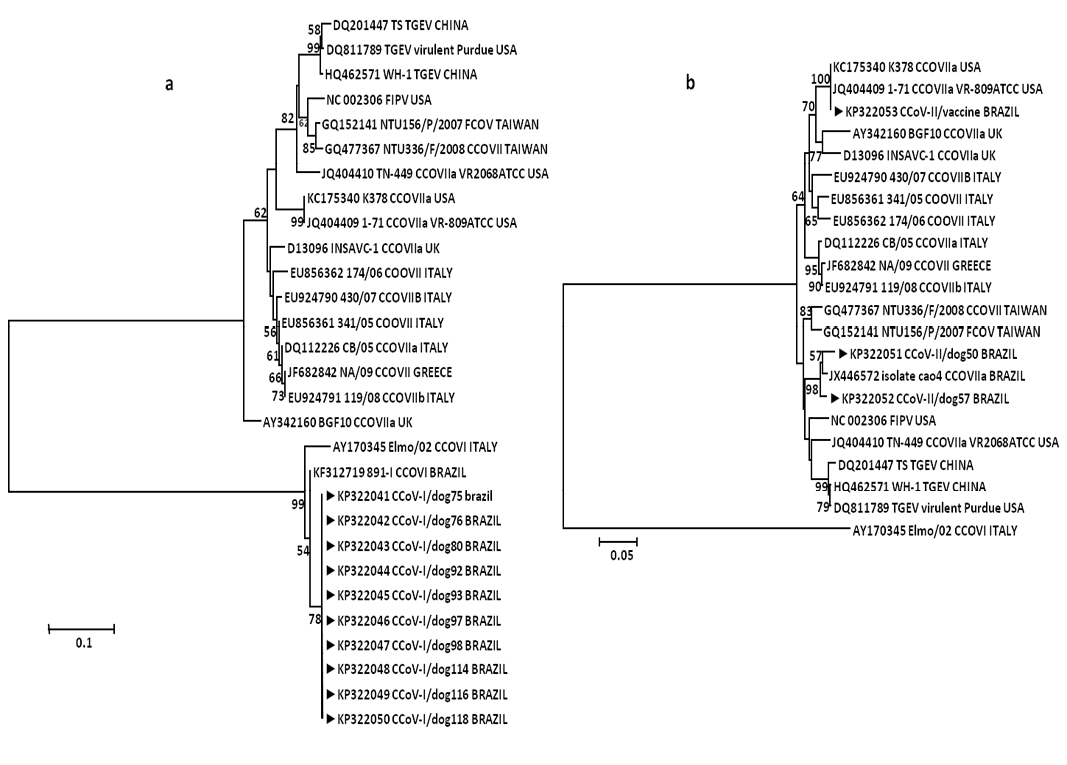
Neighbor-joining nucleotides trees (MCL model) for partial sequences of spike gene of a. CCoV type I and b. CCoV type II genes; arrows indicate strains form this study. Numbers at each node are bootstrap values (1,000 replicates). The bar represents the number of substitutions per site.