Antibiotic Resistance Coding Genes in Klebsiella pneumoniae from Clinical Cats in Bogor Indonesia
Antibiotic Resistance Coding Genes in Klebsiella pneumoniae from Clinical Cats in Bogor Indonesia
Safika1*, Ni Luh Putu Ika Mayasari1, Juliadi Ramadhan2
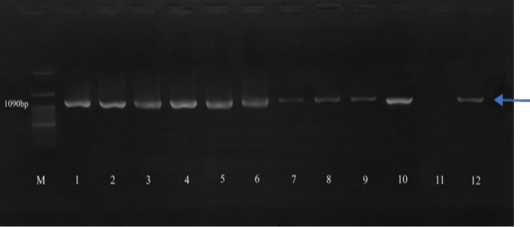
Amplification of the rpoB gene (1090 bp) in Klebsiella pneumoniae isolates. M: 100 bp DNA marker; 1 control positive ATCC 700603; 2-10 and 12 are positive for Klebsiella pneumoniae, and 11 are negative for Klebsiella pneumoniae.
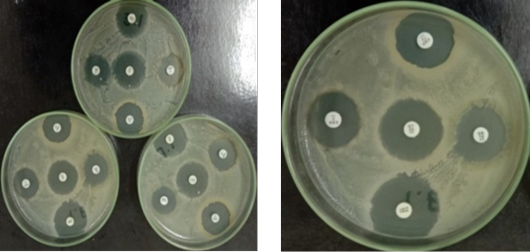
Inhibition zone of an antibiotic disc on Muller Hinton Agar.
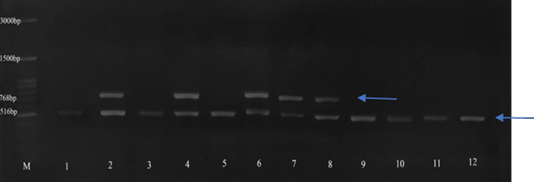
Amplification of the blaTEM (516 bp), blaSHV (768 bp) and blaCTXM (866 bp) genes. M: 100 bp DNA marker, 1 - 12: Klebsiella pneumoniae isolate that is resistant and intermediate to ampicillin antibiotics.
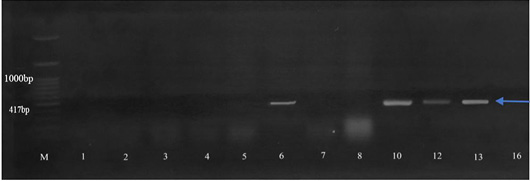
Amplification of the QnrS gene (417 bp). Isolates 6, 10, 12, and 13 were positive for QnrS gene. M: 100 bp DNA marker.
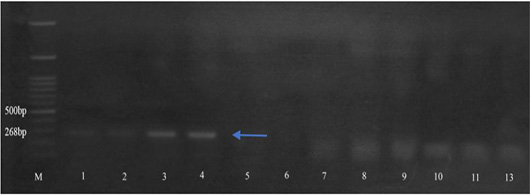
Amplification of the aac3-IV (286 bp) gene. Isolates 1-4 13 were positive for aac3-IV gene. M: 100 bp DNA marker.
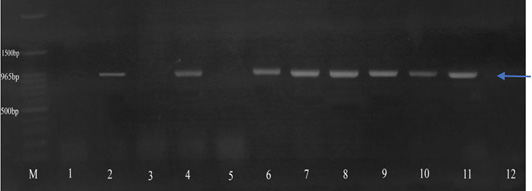
Amplification of the tetA gene (965 bp). Isolates 2, 4, 6-11 were positive for the tetA gene. M: 100 bp DNA marker.