Advances in Animal and Veterinary Sciences
Research Article
In Silico Identification of Diagnostic Candidates from Predicted Lipoproteome of Mycoplasma mycoides subsp.capri
Thachappully Remesh Arun, Rajneesh Rana*, Valsala Rekha, Thankappan Sabarinath
Division of Bacteriology and Mycology, Indian Veterinary Research Institute, Izatnagar, Bareilly, U.P., India.
Abstract | Mycoplasma mycoides subspecies capri (Mmc), the causative agent of caprine pleuro pneumonia/contagious agalactia is a major pathogen affecting goats worldwide. Development of specific immunodiagnostic assays for Mmc is often hampered by interspecies cross-reactivity with other caprine mycoplasmas and the intra species antigenic variability. The study presents a comparative and subtractive proteomic analysis to identify specific, conserved and immunogenic lipoproteins from Mmc proteome. Analysis of 896 proteins of Mmc strain 95010 predicted 72 lipoproteins by lipoP1.0 server. BLAST analysis revealed 17 putative non cross-reactive lipoproteins out of which seven were found to be conserved within the species. Further computational workflow employing ExPASy protein analysis tools and B cell epitope prediction softwares identified five lipoproteins suitable for development of immunodiagnostics.
Keywords | Mycoplasma mycoides subsp. capri, Contagious agalactia, Lipoprotein prediction
Editor | Kuldeep Dhama, Indian Veterinary Research Institute, Uttar Pradesh, India.
Received | June 12, 2017; Accepted | August 27, 2017; Published | October 08, 2017
*Correspondence | Rajneesh Rana, Division of Bacteriology and Mycology, Indian Veterinary Research Institute, Izatnagar, Bareilly, U.P., India; Email: rajneeshrana01@yahoo.com
Citation | Arun TR, Rana R, Rekha V, Sabarinath T (2017). In silico identification of diagnostic candidates from predicted lipoproteome of Mycoplasma mycoides subsp. capri. Adv. Anim. Vet. Sci. 5(10): 419-424.
DOI | http://dx.doi.org/10.17582/journal.aavs/2017/5.10.419.424
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2017 Arun et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
Mycoplasma mycoides subspecies capri (Mmc) is one of the causative agents of contagious agalactia (CA) in goats. CA is a multi-etiological syndrome caused by four different species of Mycoplasma: the other three being M. agalactiae (Ma), M. capricolum subsp. capricolum (Mcc) and M. putrefaciens (Mp). The disease is characterized by mastitis, arthritis, kerato conjunctivitis, pneumonia, septicemia and high mortality in kids (Churchward et al., 2014).The disease caused by Mmc is highly prevalent in India and is described as caprine pleuropneumonia (CPP) (Manimaran et al., 2006).
Mmc belongs to Mycoplasma mycoides cluster, which consists of five closely related mycoplasmas that cause disease in ruminants. The other four species/subspecies belonging to the cluster are M. capricolum subsp. capricolum (Mcc), M. capricolum subsp. capri pneumoniae (Mccp), M. mycoides subsp. mycoides SC (Mmm SC) and M. leachii (Ml) (Manso-Silvanet al., 2009; Thiaucourt et al., 2011). M. capricolum is a closely related species of Mmc and the Genome-To-Genome Distance (GGD) value of the pair of species is even lower than the threshold values for species delimitation (Thompson et al., 2011). The M. agalactiae shares 18% of its genome with the M. mycoides cluster as a result of frequent horizontal gene transfer events (Sirand-Pugnet et al., 2007).
Mmc strains also show considerable intra-species antigenic variability and the current Mmc serovar LC was even referred to as a separate species earlier i.e. M. mycoides subsp. mycoides Large Colony (Mmm LC) (Manso-Silvan et al., 2009; Vilei et al., 2006). The interspecies cross-reactions with other caprine mycoplasmas and intra-species variability frequently hamper the development of sensitive and specific immunodiagnostic assays (Vilei et al., 2006). Serological detection of antibodies against Mmc is generally performed using in-house ELISAs based on whole cell antigen prepared from field isolates (Assuncao et al., 2004) and there are no specific diagnostic tests available till date.
Immunoinformatics, a combination of immunology and informatics, has helped in developing methods which have been used to successfully identify antigenic epitopes in pathogens (Tomar and De, 2010). The current study employs an in silico analysis of the whole genome and proteome to identify novel diagnostic candidates from predicted lipoproteome of Mmc LC strain 95010.
MATERIALS AND METHODS
Genome/ Proteome Databases and Alignment Tools
The whole genome sequence of the Mmc strain 95010 was retrieved from the RefSeq database at the National Center for Biotechnology Information (NCBI) website (http://www.ncbi.nlm.nih.gov/) and the whole proteome was retrieved from Uniprot (Universal Protein Resource) database (http://www.uniprot.org/). All the individual nucleotide and protein sequences were retrieved and saved locally for further bioinformatics workflow as described in Figure 1.
Figure 1: Illustration of comparative and subtractive analysis workflow to identify putative diagnostic candidates in Mycoplasma mycoides subsp. capri
Lipoprotein Prediction
Lipoproteins in Mmc proteome were predicted using LipoP 1.0 server (www.cbs.dtu.dk/services/LipoP/). Peptides with predicted N-terminal cleavage sites for signal peptidase II having log-odds score greater than zero are considered as potential lipoproteins. The LipoP 1.0 algorithm produces accurate predictions of lipoproteins and discriminates between lipoprotein signal peptides, other signal peptides and n-terminal membrane helices in bacteria (Rahman et al., 2008).
Selection of Putative Non Cross Reactive Conserved Lipoproteins
All the 72 predicted lipoproteins of Mmc LC 95010 genome were subjected to protein homology searches by using the Basic Local Alignment Search Tool (BLAST) service of UniProt server (http://www.uniprot.org). BLASTp analysis was performed against the UniprotKB database using BLOSM-62 matrix with an expectation value (E) threshold of 0.0001 and filtering was applied to avoid the low complexity regions. The results were saved locally to analyze amino acid identity with other organisms and to identify the intra-species variability among the Mmc strains.
Prioritization of Identified Candidates for Heterologous Expression
Physical and chemical parameters like molecular weight, isoelectric point (pI), in vivo half-life of protein in E. coli were estimated using the protparam tool of expasy server (http://web.expasy.org/protparam/). As the members of the genera Mycoplasma utilize TGA (opal stop) codons as tryptophan coding ones, genes containing TGA codons results in premature truncation during translation in normal E. coli host cells (Minion, 1998). All the sequences were analyzed for the presence of TGA codons to identify protein targets suitable for heterologous expression in E.coli.
B Cell Epitope Analysis and Antigenicity Prediction Using Vaxijen Server
Selected candidates were further analyzed by in silico antigenicity prediction server, VaxiJen (http://www.jenner.ac.uk/VaxiJen). It is the first server developed for alignment-independent prediction of protective antigens of bacterial, viral and tumour origin (Doytchinova et al., 2007). Immunogenicity of the proteins was also evaluated using Bepipred B cell epitope prediction software (http://tools.iedb.org/bcell/). The method combines the hidden Markov model with propensity scale methods for predicting linear B-cell epitopes (Larsen et al., 2006). The results were used to prioritize the selected proteins and those with significant B cell epitopes were selected so that they can be expressed as recombinant proteins in a heterologous ex pression system.
Table 1: Details of five putative lipoproteins identified as diagnostic candidates in Mycoplasma mycoides subsp capri
|
SI No. |
Uniprot protein ID |
Gene ID |
Amino acid length |
Number of TGA codons with position |
pI value |
Molecular weight (kDa) |
Best BLASTp hit outside the species: Organism, percent identity |
BLASTp within the species: Mmc strain, percent identity |
| 1 | F4MNX4 | MLC_0780 | 466 | No TGA codon | 5.35 | 52.23 |
Mcc1, 34% |
Mmc strain PG3, 91% |
| 2 | F4MNX7 | MLC_0810 | 563 | 4 (112, 265, 378, 489) | 5.34 | 63.34 |
Mcc, 46% |
Mmc strain PG3, 93% |
| 3 | F4MNX8 | MLC_0820 | 790 | 5 (97, 165,166, 206, 382,483) | 5.27 | 90.79 |
Ma2, 28% |
Mmc strain PG3, 94% |
|
4 |
F4MPK9 | MLC_3130 | 172 | 2 (113, 165) | 8.88 | 18.19 |
Mcc, 42% |
Mmc strain PG3, 95% |
| 5 | F4MR59 | MLC_8630 | 247 | 5 (97, 130, 214, 231, 238) | 5.07 | 25.39 |
Mfer3, 40.4%
|
Mmc strain PG3, 91.5% |
1M. capricolum subsp. capricolum (Mcc); 2M. agalactiae (Ma); 3M. ferriruminatoris (Mfer)
RESULTS
Sequences and Databases
The Mmc LC 95010 has a circular chromosome of 1,153,998 bp (GenBank accession number NC_015431) which consists of 922 putative CDS and a plasmid coding for 2 proteins (Thiaucourt et al., 2011). The whole proteome of Mmc (Proteome IDUP000010103) consists of 896 proteins which were mapped to 921 gene IDs. The putative CDS MLC_7780 was not found in the proteome as it was a pseudo gene and 19 proteins were identified to be coded by multiple genes.
Lipoprotein Prediction
Analysis of 896 proteins of Mmc LC 95010 using the LipoP 1.0 identified 72 proteins with predicted cleavage sites for signal peptidase II and 73 proteins with a predicted cleavage site for signal peptidase I. All the 72 lipoproteins with cleavage sites for signal peptidase II were selected for further bio informatic analysis and diagnostic target prioritization.
Blast Analysis
Lipoproteins which showed less than 50% identity with the other organisms were considered as putative non cross reactive proteins of Mmc. BLAST analysis revealed 17 putative lipoproteins which qualified the selection criterion. The non-cross reactive proteins were further analyzed for intra-species variation. Out of 17 putative lipoproteins, only 7 candidates were having more than 90% identity with the other Mmc strains PG3 and GM12. The absence of a hit against Mmc strain GM12 was not considered as a criterion for exclusion, since only the partial proteome (372 proteins) is available for Mmc strain GM12.A complete proteome with 779 protein entries was available for Mmc type strain PG3 and all the lipoproteins having a homolog with more than 90% identity in the strain were considered to be conserved.
Antigenicity Prediction Using Vaxijen Server and Bepipred Epitope Prediction Tool
For antigenicity prediction using the VaxiJen server, a threshold value of 0.5 was selected and the candidates which gave a value above cut-off were considered as predicted antigens. Analysis of 7 selected putative lipoprotein predicted five to be antigenic, whereas two proteins -Uniprot IDsF4MPK7 (MLC_3110) and F4MPS5 (MLC_3800) were predicted to be non antigenic. Bepitope linear B cell epitope prediction server identified epitopes in all the five selected lipoproteins - F4MNX4 (MLC_0780), F4MNX7 (MLC_0810), F4MNX8 (MLC_0820), F4MPK9 (MLC_3130) and F4MR59 (MLC_8630) as given in Figure 2.
Prioritization of Diagnostic Candidates Based on Protein Parameters
Presence of TGA stop codons was identified and protein parameters- molecular weight, isoelectric point (pI) and in vivo half life of protein in E. coli were estimated for the selected five conserved, non cross reactive lipoproteins. There was only one lipoprotein (Uniprot IDs F4MNX4 without any TGA codons in the entire length of the gene sequence. The other candidates have immunogenic regions without TGA codons that can be expressed in prokaryotic expression system for the development of sero diagnostics. The details of bio informatic analysis of selected diagnostic candidates are given in Table 1.
There was an additional set of four candidates (F4MP11, F4MQW4, F4MQW9 and F4MRA1) which have cross reactivity only with the closely related bovine pathogens. M. mycoides subsp. mycoides SC, M. leachii and wildlife pathogen M. ferriruminatoris These can also be evaluated in diagnosis of Mmc as the above mentioned organisms are not caprine pathogens.
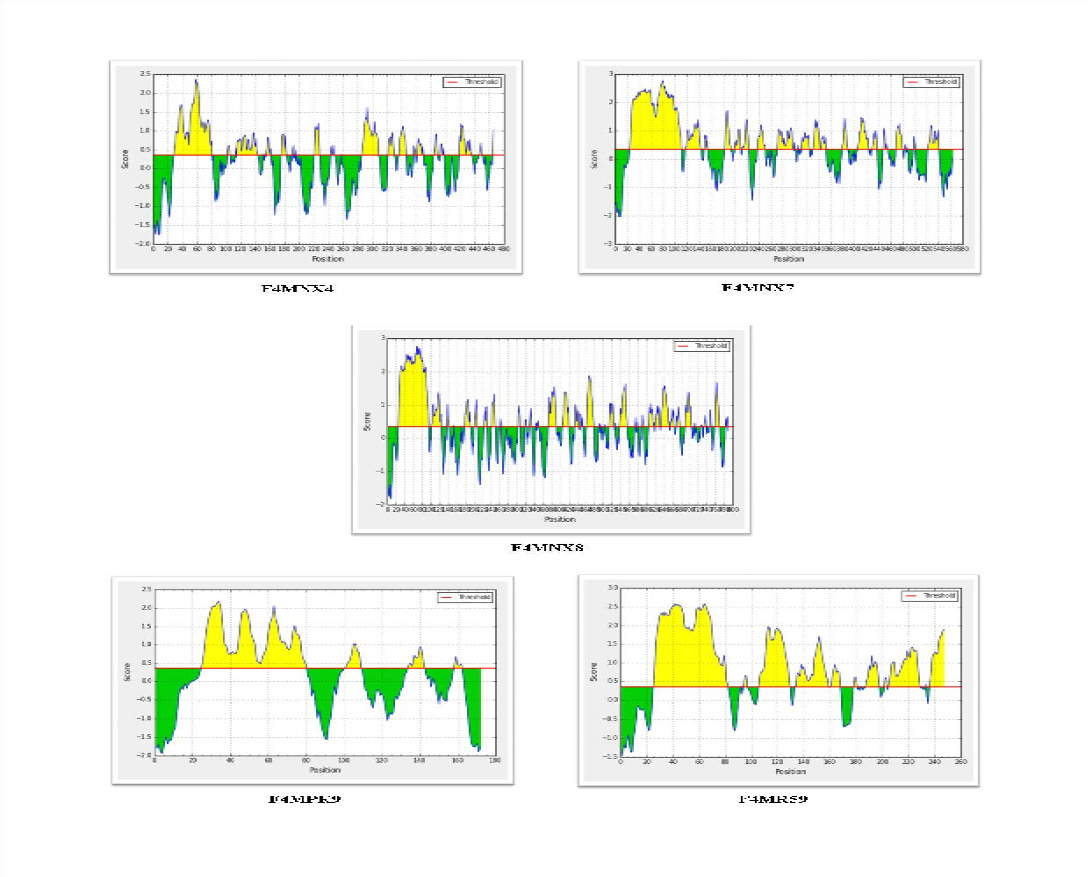
Figure 2: Prediction of linear B cell epitopes in the five selected lipoproteins using Bepipred server. The regions above the threshold (0.35) are antigenic, shown in yellow while, green color reflects the polypeptide regions that could not satisfy the threshold margin.
DISCUSSION
Mycoplasmal lipoproteins are excellent immunogens, which have been used for sero diagnosis of various Mycoplasma spp. (Bruderer et al., 2002; Fusco et al., 2007; Alberti et al., 2008). Hence, the identification of conserved immunogenic lipoproteins in Mmc is an important step towards development of sensitive and specific immunodiagnostics. Churchward et al. (2014) performed immuno proteomic characterization of Mmc by mass spectrometry analysis two-dimensional (2D) electrophoresis spots and western blot. The proteins identified in these studies were mostly metabolic enzymes and other cytoplasmic proteins. Although the identified immunogens can be good vaccine candidates, their utility as diagnostic candidates was limited because of the high similarity with closely related mycoplasmas. Even the 2D electrophoresis and western blot analysis of liposoluble proteome (Corona et al., 2013) could not identify any lipoproteins. The lack of lipoproteins and other membrane-associated proteins identified in these studies is probably due to their low abundance in comparison to other cellular proteins and lack of solubility when preparing the samples for isoelectric focusing (Churchward et al., 2014).
In the current study, putative diagnostic candidates were identified using a bio informatic workflow employing lipoprotein sequence prediction, BLAST analysis, Vaxigen antigenicity prediction server, Bepipred linear epitope prediction tool and protein parameters including presence of TGA codons. Our study identified 72 putative lipoproteins using LipoP 1.0 server which has been used earlier for lipoprotein identification from other Mycoplasma spp. including the closely related M. mycoides subsp. mycoides SC (Heller et al., 2016).
BLASTp analysis was performed to identify the putative non cross reactive lipoproteins in Mmc proteome. Due to the high genetic similarity within mycoides cluster, there were no proteins with “no hits” in the BLASTp analysis. Most investigators describe protein similarity in terms of “percent identity” of amino acids whereas E-values and bit-scores are also useful for inferring homology. Non-cross-reacting domains usually show less than 70% sequence identity, emphasizing the genetic basis for immunological specificity (Maeland et al., 2015). McNulty et al. (2015) adopted the criterion of 70% amino acid sequence identity over more than 70% of the total protein length for identification of putative diagnostic antigens from Onchocerca volvulus. Their study identified 60 diagnostic candidates, which satisfy the criterion from a total of 241 immunoreactive proteins analyzed. Here, we adopted a more stringent criterion of less than 50% identity over the entire length of protein to identify 17 non cross-reactive ones out of 72 lipoproteins. When two protein sequences have less than 50% identity, the risk of cross reactivity is expected to be rare (Silvanovich et al., 2006). Although it is difficult to predict antibody cross-reactivity based on global sequence similarity, this level of conservation makes these proteins more attractive immunodiagnostic candidates than those having orthologues in related species.
Significant protein variability within the species is well documented for various Mycoplasma spp. (Calus et al., 2007; Salam et al., 2013). Fischer et al. (2012) conducted a multi locus sequence typing (MLST) analysis of 33 Mmc isolates and identified a very high genetic diversity within the species. The putative diagnostic candidate for Mmc should also address the high intra-species variability within Mmc. In our study, seven out of 17 non cross reactive lipoproteins showing 90% amino acid identity over the entire length of protein were selected for further evaluation.
VaxiJen is the first server for alignment-independent prediction of protective antigens. It allows antigen classification based on the physicochemical properties of proteins without depending on the sequence alignment. It has been used for prediction of vaccine candidates in several bacterial species including M. agalactiae (Forouharmehr and Nassiry, 2015). Five candidates which were identified to be antigenic by VaxiJen server can also be used as potential vaccine candidates for Mmc. All the identified targets MLC_0780, MLC_0810, MLC_0820, MLC_3130 and MLC_8630 are uncharacterized proteins. Recently, an in silico analysis combined with 2D electrophoresis and western blot predicted eight novel uncharacterized antigens to have high immunological value and Mbov_0579 was found to be the best antigenic target for sero diagnosis of M. bovis (Khan et al., 2016).
This study identified novel diagnostic candidates, which can be utilized in the development of sensitive and specific diagnostic tests and recombinant vaccines against Mmc. Our selection procedure theoretically guarantees that the identified lipoproteins have the potential to achieve an adequate degree of specificity and sensitivity, minimizing the likelihood of false positives associated with current diagnostic tests. The predicted lipoproteins need to be expressed in suitable prokaryotic expression system and validated for the development of immunodiagnostics.
ACKNOWLEDGEMENTS
We thank Director, Indian Veterinary Research Institute for funding the necessary facilities for the research programme under which the current study is conducted. The first author is thankful to Department of Science and Technology (DST), India for providing financial support (Inspire fellowship) during the period this research work.
CONFLICTS OF INTEREST
The authors declare that there is no conflict of interest.
authors contribution
Thachappully Remesh Arun and Valsala Rekha performed the in silico analysis work. Rajneesh Rana supervised the work and aided in writing the manuscript. Thankappan Sabarinath aided in writing the manuscript.
REFERENCES






