Advances in Animal and Veterinary Sciences
Review Article
Peste-Des-Petits-Ruminants: An Indian Perspective
Dhanavelu Muthuchelvan1*, Kaushal Kishor Rajak1, Muthannan Andavar Ramakrishnan1, Dheeraj Choudhary1, Sakshi Bhadouriya1, Paramasivam Saravanan2, Awadh Bihari Pandey1, Raj Kumar Singh3
1Division of Virology, Indian Veterinary Research Institute, Mukteswar Campus, Nainital, Uttarakhand 263 138, India; 2Indian Veterinary Research Institute, Hebbal Bengaluru, 560024, Karnataka, India; 3Indian Veterinary Research Institute, Izatnagar, 243122, India.
Abstract | Peste-des-petits-ruminants (PPR) is an acute or subacute, highly contagious viral disease of small ruminants, characterized by fever, oculonasal discharges, stomatitis, diarrhoea and pneumonia with high morbidity and mortality. Peste-des-petits-ruminants virus (PPRV), the etiological agent of PPR, is antigenically related to another rinderpest virus (RP) which was globally eradicated. PPR is gaining worldwide attention through the concerted effort of scientists working together under the aegis of global PPR research alliance (GPRA). The first homologous live attenuated vaccine was developed using Nigeria 75/1, which has been used worldwide. In India, live attenuated vaccines have been developed using Sungri 96, Arasur 87 and Coimbatore 97 viruses. In this review, the status of PPR and control strategy with special reference to the Indian context is comprehensively discussed.
Keywords | PPR, PPRV, Vaccine, DIVA, Eradication, Symptoms, Epidemiology, Diagnosis, Vaccines, Immunity, Control programme, Replication
Editor | Muhammad Munir (DVM, PhD), Avian Viral Diseases Program, Compton Laboratory, Newbury, Berkshire, RG20 7NN, UK.
Received | April 27, 2015; Revised | June 16, 2015; Accepted | June 18, 2015; Published | June 24, 2015
*Correspondence | Dhanavelu Muthuchelvan, Indian Veterinary Research Institute, Nainital, Uttarakhand, India; Email: drchelva@gmail.com
Citation | Muthuchelvan D, Rajak KK, Ramakrishnan MA, Choudhary D, Bhadouriya S, Saravanan P, Pandey AB, Singh RK (2015). Peste-des-petits-ruminants: An Indian perspective. Adv. Anim. Vet. Sci. 3(8): 422-429.
DOI | http://dx.doi.org/10.14737/journal.aavs/2015/3.8.422.429
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2015 Muthuchelvan et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
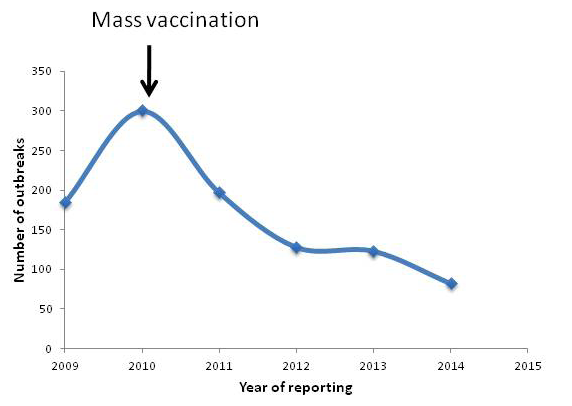
Peste-des-petits-ruminants (PPR) is a viral disease of sheep and goats caused by PPR virus (PPRV) with huge economical concern. India harbours 65.06 and 135.17 million of sheep and goats, respectively (19th Livestock census, 2012; http://dahd.nic.in/dahd/WriteReadData/Livestock.pdf) which is 16.1 and 6.4 % of the world’s total goat and sheep population, respectively. The disease threatens 70% of the landless labourers, small and medium farmers who rears sheep and goats for their livelihood. As the disease affects the poor sections of the society, international agencies such as FAO & OIE set a target for global eradication by 2030 (http://www.fao.org/ppr/en/). The disease causes up to 100% mortality in and productive loss. It is estimated that PPR alone leads to annual losses of 1,800 million Indian Rupees (US$ 39 million) (Singh, 2012). As one of the forerunners in the rinderpest (RP) research and significant contribution in the global eradication of the disease, consequently, our lab developed live attenuated vaccine (Sungri96) and monoclonal antibody based diagnostics (Singh et al., 2004b, c). The vaccine and diagnostics are supplied throughout India as well as neighbouring countries. Availability of these technologies helped India to initiate mass vaccination campaign which brought down the incidence of the disease significantly (Figure 1). Further, Government of India has recently launched a Peste des Petits Ruminants Control Programme (PPR-CP). The PPR Control Programme involving intensive vaccination of susceptible animals has been started in 2010. The programme involves vaccinating all susceptible goats and sheep and three subsequent generations. Under first phase, States of Kerala, Tamil Nadu, Karnataka, Andhra Pradesh, Maharashtra, Goa and UTs of Lakshadweep, Daman and Diu, Dadra and Nagar Haveli, Andaman and Nicobar Islands and Pondicherry were covered. In the second phase, the programme has been expanded to all States/UTs in February, 2014 http://dahd.nic.in/dahd/WriteReadData/Animal%20Husbandry%20English%202014-15%20(1).pdf. This short review focuses on the current scenario of PPR in India.
The Virus
Peste-des-petits-ruminants virus (PPRV) is a member of the genus Morbillivirus. subfamily Paramyxovirinae, family Paramyxoviridae in the order Mononegavirales (International Committee on Taxonomy of Viruses, 2012). Since the virus is enveloped one, it is easily inactivated under sunlight and with many chemicals. The genome of PPRV is a negative sense, single-stranded RNA with the size of 15948 bp. The genome is organized into six transcriptional units and each encodes at least one non overlapping protein: the nucleocapsid (N), the matrix (M), the polymerase or large (L), the phosphoprotein (P), and two envelope proteins haemagglutin (H) and fusion (F). The P protein uses alternate expression strategies to code for two non-structural proteins viz., V and C (Muthuchelvan et al., 2006). The PPRV genes arranged from 3ˈ to 5’ on the genome is in an order of N-P-M-F-H-L separated by intergenic region which is CTT in most cases. At the 3’ and 5ˈ end of the genome there is a leader (52 nucleotides) and trailer (37 nucleotides) region contain promoter functions (Bailey et al., 2005). Two heptad repeats (HR1 and HR2) of the fusion protein was shown to form a six-helix and trimeric coiled-coil bundle for initiation of fusion activity (Rahaman et al., 2003). Host-pathogen interaction study with vaccine virus (Sungri96) and goat peripheral blood mononuclear cells (PBMCs) revealed the dysregulation of immune regulatory pathways (Manjunath et al., 2015). Establishment of an in vitro system using Sungri96 virus shown that RNP complex is actively synthesis mRNA (Yunus and Shaila, 2012). PPRV multiplication was reduced significantly in Signaling Lymphocyte Activation Molecule (SLAM;CD150) receptors suppressed with siRNA in B95a (Pawar et al., 2008b).Transiently expressed PPRV F glycoprotein induces cell fusion in the absence of H protein (Seth and Shaila, 2001). The H protein was found to have potential T cell determinant(s) at amino acids position 123-137 and 242-609 (Sinnathamby et al., 2001).
The disease
PPR is a disease of small ruminants which resembles rinderpest of cattle. The incubation period is 3-6 days, followed by high fever, oculonasal discharges, pneumonia, stomatitis, and inflammation of gastrointestinal tract leading to severe diarrhoea followed by death or recovery (Balamurugan et al., 2014a; Sen et al., 2010; Zahur et al., 2008). The disease causes more severe lesions in goats than sheep. Although, the reason for this host specificity is not fully understood, difference in genetic makeup and/or receptor distributions of the host might have a role. Different levels of SLAM mRNA could influence the virus replication in different species (Pawar et al., 2008a). Another study examined the replication of PPRV in PBMCs of Indian goats and water buffalo and demonstrated that the level of TLR3 and TLR7 and downstream signalling molecules correlate with susceptibility (Dhanasekaran et al., 2014). Incidence in other species such as cattle, pig, camel, buffalo, lion and captive wild small ruminants were reported but are not contributing to the disease epidemiology. Experimental infection of cattle with PPRV revealed that the nucleic acid could be detected up to 397 days post infection (Sen et al., 2014).
No carrier state or persistent infections have been reported, however, disease may circulate in subclinical forms in sheep and goats of endemic region. The recovered animals are immune for lifelong. The cytokine expression profile (IL-4 and IFN-γ) of vaccinated and infected goats were found to have a unique biphasic response of IL-4 expression with up-regulation of IFN-γ on 7th days post vaccination (Patel et al., 2012). PPRV was reported to induce apoptosis in vitro in goat PBMCs (Mondal et al., 2001).
Although, presence of PPR-like disease has been suspected earlier in retrospective study (Taylor et al., 2002), its presence was confirmed in India in1987 from Arasur village of Villupuram district of Tamil Nadu (Shaila et al., 1989). Currently, PPR outbreaks are being reported regularly from different parts of the country (Chauhan et al., 2011; Kerur et al., 2008; Muthuchelvan et al., 2014; Nanda et al., 1996; Raghavendra et al., 2008; Singh et al., 2004a). The disease outbreak status for the period between 2009 and 2014 is depicted in Figure 1. The reported seroprevalence rate of PPRV at country level in goats and sheep was 43.56 % (Balamurugan et al., 2011) and 4.58% in cattle and buffalos (Balamurugan et al., 2012a). Molecular epidemiological studies confirm the circulation of lineage IV in India (Balamurugan et al., 2010; Dhar et al., 2002; Muthuchelvan et al., 2014; Shaila et al., 1996). Presence of mixed infection with bluetongue (Mondal et al., 2009), goatpox (Malik et al., 2011) and Orf (Saravanan et al., 2007) viruses have also been reported.
Disease transmission and Epidemiology
The virus spreads through close contact between infected and susceptible population. The primary portal of entry is via respiratory route. Viraemia develop 2-3 days post infection i.e 1-2 days before the appearance of first clinical sign. Fine infective droplets from the secretions and excretions of the infected animals are released into the air (Sen et al., 2010). Transmission could also occur through contaminated water, feed troughs and bedding (Lefèvre and Diallo, 1990).
In India, animals are allowed to share common grazing land and water sources. Besides, migration of animals between various states is common especially, in the sub-Himalayan region and western dry land areas such as Rajasthan and Gujarat (Nanda et al., 1996; Singh et al., 2004a). Mixing of these migrated populations with local population may contribute to the disease transmission. Further, during the festival seasons, animals are shipped to various states for meat purposes. These unrestricted movements of animals contribute significantly to the epidemiology of the disease. In India, the disease occurs round the year and the maximum outbreaks reported during the winter and rainy seasons. Therefore, vaccination just prior to the onset of rainy/winter season will be more appropriate.
Initially, PPRV was classified in to 4 lineages I–IV based on the F gene sequencing (Dhar et al., 2002; Shaila et al., 1996); lineage I-III viruses were reported in several countries of Africa and lineage IV (Asian lineage) mainly in Middle East and Asia (Banyard et al., 2010; Dhar et al., 2002; Ozkul et al., 2002). Currently, N gene is preferred over F gene due to its better molecular separation (Kwiatek et al., 2007). In the recent past, many researchers reports the presence of lineage IV in African countries (Maganga et al., 2013; Salami et al., 2014). Till now, only lineage IV viruses have been reported in India. The PPRV goat strain isolated during the recent outbreak at Tripura showed 99.2 to 99.6% nucleotide identities with the Bangladesh strains (Muthuchelvan et al., 2014). The latter study confirms the transboundary transmission of PPRV with the neighbouring countries.
Vaccine
Before the development of PPRV vaccine, owing to cross-protection between RPV and PPRV, an attenuated tissue culture RP vaccine (TCRP; Plowright’s strain) was used in many countries. However, since the last phase of RP eradication, this practice was banned. The first homologous vaccine against PPR was developed in 1987 using a Nigerian isolate (Nigeria/75/1) (Diallo et al., 1989, 2007). Subsequently, three PPRV vaccines have been developed in India using indigenous isolates of lineage IV (PPRV/Sungri/96, PPRV/Arasur/87, and PPRV/Coimbatore/97) (Palaniswami et al., 2005; Singh et al., 2004a). Although, all the three vaccines were reported to be equally efficacious (Santhosh et al., 2013; Saravanan et al. 2010), it was observed that the replication cycle and monoclonal reactivity of Arasur/87 virus is different from Sungri/96 virus (Singh et al., 2010). Currently, Sungri/96 strain is being used throughout the country. This vaccine is potent, confers life-long immunity and is safer to pregnant animals (Rajak et al., 2005). Cold chain maintenance is the major concern of live attenuated vaccines in tropical countries. Attempts were made to develop thermal adapted vaccine by growing the virus at high temperature (>40°C) (Balamurugan et al., 2014b; Riyesh et al., 2011). Being a live attenuated vaccine, animals mount strong immune response against all the viral proteins and therefore differentiating the infected from the vaccinated animals (DIVA) would be difficult. Developing genetically marked vaccines would be advantageous during the eradication phase. Subunit vaccines prepared by expressing the PPRV F and/or H gene in pox and adeno viral vectors found to be experimentally protective (Caufour et al., 2014; Chandran et al., 2010; Chen et al., 2010; Diallo et al., 2007; Herbert et al., 2014; Rojas et al., 2014). Attempt was made to develop a subunit vaccine by expressing the H protein of PPRV in peanut plants (Arachis hypogea) and found to elicit neutralizing antibody responses in sheep (Khandelwal et al., 2011). The major disadvantage of this strategy is need of multiple doses for protective immunity. Another approach is to manipulate a specific region or epitope of a viral protein of the existing vaccine to obtain positively and or negatively marked vaccines. Recently, two groups succeeded in developing DIVA vaccines using Nigeria 75/1 strain (Hu et al., 2012; Muniraju et al., 2015). Our lab is currently working on development of a DIVA vaccine for PPRV Sungri/96. Experimental combined vaccines against PPR/goatpox (Hosamani et al., 2006), and PPR/sheeppox (Chaudhary et al., 2009) have been shown as promising candidates. Appropriate period of vaccination in kids was found to be four months (Balamurugan et al., 2012b).
Diagnostics of PPR
Laboratory confirmation of the disease is usually done through virus isolation and virus neutralization assay, which are time consuming and laborious. Several assays have been described to detect virus-specific antibodies or viral antigens.
Detection of Antibodies Against PPRV
A monoclonal antibody based competitive ELISA (cELISA) for detection of antibodies against PPRV has been developed in our laboratory (Singh et al., 2004a). The assay was compared with VNT and found to be 92.2% and 98.84%, of specificity and sensitivity, respectively. Although, this test is suitable for mass screening, it uses live virus as positive antigen, which could be a disadvantage for use in PPR free countries/regions. To overcome this, we are currently working on a recombinant antigen based competitive ELISA. Similarly, a polyclonal antibody based indirect ELISA was developed for detection of antibodies against PPRV in the serum samples (Balamurugan et al., 2007). The performance of the assay was comparable with that of c-ELISAs.
Detection of PPRV Antigen
Virus Isolation
Virus isolation remains the “gold standard” for the diagnosis of PPR. Blood collected at the height of the temperature is the best material for this purpose. The nasal or ocular swabs or 10% tissue suspension can also be used. The PPRV can be propagated in vitro in several primary bovine and sheep cells, as well as established cell lines like Vero (African green monkey kidney cells) and B95a (Marmoset B-lymphoblastoid cells) (Sreenivasa et al., 2006). In most of these cells, the PPRV manifests morbillivirus-specific CPE by 3-5 days post infection. In some cases, up to five blind passages are needed for isolating the virus.
Sandwich ELISA
The sandwich-ELISA for the detection of PPRV antigen in the clinical samples was developed at our laboratory. This assay uses a monoclonal antibody directed against N protein. The test is compared with IC-ELISA (BDSL) and found to have 89% and 93%, sensitivity and specificity, respectively (Singh et al., 2004b). This assay is easy to perform and is routinely adopted by many diagnostic laboratories. Like c-ELISA, in this assay also live attenuated PPRV used as positive antigen. Attempts were made to develop a recombinant N protein based ELISA (Yadav et al., 2009).
Detection of PPRV Nucleic Acid
Reverse Transcription-Polymerase Chain Reaction (RT-PCR) is the method of choice for detecting nucleic acids of PPRV in clinical samples. RT-PCRs have been reported for detection and differential diagnosis of RP and PPR viruses targeting the N and F gene (Balamurugan et al., 2006; Couacy-Hymann et al., 2002; Forsyth and Barrett, 1995). N-Gene based PCR-ELISA has developed at IVRI, Mukteswar to detect PPRV from clinical samples. The test can detect viral RNA in infected tissue culture fluid with a titre as low as 0.01 TCID50 and is used to evaluate only critical samples (Saravanan et al., 2004). PCR based on other genes also are available (Brindha et al., 2001; George et al., 2006).
Real-time RT-PCR and Lateral Flow Assays
Real time RT- PCR has been used for quantification and diagnosis of PPR virus. M gene-based hydrolysis probe (TaqMan), SYBR Green I based real-time RT-PCR assays targeting the M gene of PPRV are in use (Abera and Thangavelu, 2014; Balamurugan et al., 2012c). Recently, a novel non-amplification technique was developed to detect nucleic acids of PPRV in which two probes complementary to the target sequences - one conjugated to magnetic microparticles and the second to gold nanoparticles labelled with horseradish peroxidase (Tao et al., 2013). The assay has great potential due to quick performance (45 min) and not requiring expensive instrumentations etc.
Recently, an immunochromatographic test has been developed for the diagnosis PPR under field conditions. The assay has been validated with clinical samples collected in Ivory Coast, Pakistan, Ethiopia and Uganda (Baron et al., 2014).
Loop Mediated Isothermal Amplification (LAMP) Assay
LAMP assay for rapid detection of PPRV from clinical samples has been developed using M gene (Li et al., 2010). Similar assay has been developed in our laboratory using N gene and the assay is 100 and 1000 times more sensitive than RT-PCR and s-ELISA, respectively (Dadas et al., 2012).
Control program in India
Focused vaccinations in high-risk populations and mass vaccination are necessary to achieve 70 to 80% of herd immunity. The control strategies should follow the “bottom-up” approach (farmers to field veterinarians to policy makers) (Singh, 2011). Besides, strengthening of PPR vaccine production and quality control units, disease diagnostic laboratories, infrastructure for field Veterinarians are also key factors. The NCP-PPR control programme started in 2010 and currently expanded to all the provinces of the country.
Conflict of interest
There exists no conflict of interest.
Acknowledgements
The authors thank ICAR-Indian Veterinary Research Institute for necessary support.
Author’s cONTRIBUTION
Dhanavelu Muthuchelvan, Dheeraj Choudhary, Kaushal Kishor Rajak, Sakshi Bhadouriya and Paramasivam Saravanan collected the literature. Dhanavelu Muthuchelvan wrote the manuscript. Dhanavelu Muthuchelvan, Kaushal Kishor Rajak and Muthannan Andavar Ramakrishnan edited the manuscript. Awadh Bihari Pandey and Raj Kumar Singh performed the final check.
References