Advances in Animal and Veterinary Sciences
Research Article
Insight into Isolation, Identification and Antimicrobial Sensitivity of some Moulds Isolated from Fresh Water Fishes
Ashraf A. Abd El Tawab1, Fatma I. El Hofy1, Eman M. Moustafa2*, Marwa R. Halawa3
¹Bacteriology, Immunology and Mycology Department, Faculty of Veterinary Medicine, Benha University, Egypt; 2Department of Fish Diseases and Management, Faculty of Veterinary Medicine, Kafr El-Sheikh University, Kafr El-Sheikh governorate, Postal code: 33516, Egypt; 3Central Diagnostic and Research Lab, Faculty of Veterinary Medicine, Kafrelsheikh University, Egypt.
Abstract | The expanding recurrence of invasive fungal infection and the high mortality associated with dispersed fungal disease have highlighted the requirement for fast recognizable proof of infectious molds. The current study was conducted to identify the most prevalent mycotic diseases among cultured freshwater fishes with special focus on isolation, molecular identification and antimicrobial sensitivity testing. A total number of 300 examined cultured freshwater fishes; 200 Oreochromis niloticus (120 apparently healthy and 80 showing clinical signs) and 100 Clarias gariepinus (30 apparently healthy and 70 showing clinical signs), were collected alive from five freshwater fish farms at Kafr El Sheikh Governorate all over the year 2018. Clinically, examined Oreochromis niloticus revealed scale detachment, fins erosion, pale body coloration and eye opacity particularly in fish naturally infected with Aphanomycess, Aspergillus, Pencillium and Fusarium; however, infected Clarias gariepinus showed scattered hemorrhagic patches on the ventral abdomen. Mycological examination revealed the isolation of 900 fungal isolates; 570 fungal isolates from Oreochromis niloticus and 330 isolates from C. gariepinus. Aphanomyces invadans was the predominant fungi among diseased fishes mainly from skin, fins and gills. However, Aspergillus flavus, Aspergillus niger and Alternaria sp. were the most predominant fungi isolated from apparently healthy fishes mainly from internal organs. Molecular identification was performed using inter-transcriped spacer (ITS) gene for the most prevalent isolates among either diseased or apparently healthy fishes. Developed PCR assay specific primer detect not only Aphanomyces invadans showing clear bands at 569 bp molecular weight, but also Asperigillus flavus and Asperigillus niger showing clear bands at 595 and 600 bp molecular weight, respectively. Anti-fungal sensitivity test revealed that all mould isolates are sensitive to Nystatin and the majority was resistant to Voriconazole. Briefly, being normal mycoflora, most fungi still can cause infections to fishes. Consequently, proper health management practices should be adapted while fish rearing.
Keywords | Clarias gariepinus, Fungi, Identification, Oreochromis niloticus, PCR
Received | November 02, 2019; Accepted | January 10, 2020; Published | February 20, 2020
*Correspondence | Eman Moustafa Moustafa, Department of Fish Diseases and Management, Faculty of Veterinary Medicine, Kafr El-Sheikh University, Kafr El-Sheikh governorate, Postal code: 33516, Egypt; Email: emantarek2002@yahoo.com
Citation | El-Tawab AAA, El-Hofy FI, Moustafa EM, Halawa MR (2020). Insight into isolation, identification and antimicrobial sensitivity of some moulds isolated from fresh water fishes. Adv. Anim. Vet. Sci. 8(2): 174-182.
DOI | http://dx.doi.org/10.17582/journal.aavs/2020/8.2.174.182
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2020 El-Tawab et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
Fish represents the most important source of human dietary protein worldwide, especially in African countries (Hussain et al., 2011; Rubbani et al., 2011). Natural fisheries are dynamically exhausted and the aquaculture wind up imperative intends to compensate the lack; providing a valuable and fundamental protein source for human consumption (Fletcher et al., 1999; Dawood and Koshio, 2019). Aquaculture contributes more than half of the total fish production in the world and about 77% of the total Egyptian fish production (Subasinghe and Soto, 2009; Mur, 2014).
Nile Tilapia (Oreochromis niloticus) and African catfish (Clarias gariepinus) are the most prevalent freshwater fishes in Egypt because of their rapid growth rate with excellent meat quality, market demand, resistance to diseases and ability to withstand adverse environmental conditions (Goda et al., 2007; Amin et al., 2019; Dawood et al., 2019a, b).
The intensification and extension of the aquaculture industry are subjected to several ecological stressors (Awad and Awaad, 2017). It was accounted that disease is a final product of a complex collaboration of the host, its environment and the pathogen itself where the environmental stress is viewed as a vital inciting element regarded as a crucial inducing factor causing various diseases (Duan et al., 2017; El-Deep et al., 2019; Vicente et al., 2019). Adverse aquaculture conditions can enhance fish diseases transmission, especially infectious fungal diseases, resulting in limited fish production and subsequent financial losses (Faruk et al., 2004; Rodolphe et al., 2014; Ashour et al., 2017).
The importance of fungal diseases in freshwater fish not stopped just for excessive mortalities but also as economic issue, such as growth rate depletion, decreased hatchability especially in chronic infection or by mycotoxins creation by polluted organism in case of bad stocking feed. Fungal infections in fish are viewed as secondary cause of disease; fungi affecting fishes are opportunistic and attack the fishes only if they are stressed or immune-compromised. Primary causes may be attributed to temperature change, water quality problems, poor conditions, skin injury as a result of rough handling, bacterial disease and/or parasites (Verma, 2008; Chauhan et al., 2014; Noor El-Deen et al., 2018). In spite of the fungal infections importance, the available knowledge about them is still poor for two basic causes: difficult identification of pathogenic fungi and the growth of saprophytic organisms once the fish is dead (Bassiouny et al., 2019).
The most common widely recognized fungal infection was the pathogenic oomycetes; which belong to the order Saprolegniales, including the genera Saprolegnia, Achlya and Aphanomyces. Saprolegniosis has significant impact on freshwater ecosystems by infecting different host organisms and their eggs (Kiesecker et al., 2001; Fernández-Benéitez et al. 2008; Ruthig 2009; Eissa et al., 2013). Some species of Aphanomyces invadans are regularly in charge of serious disease outbreaks in fish farms and in the common habitat causing enormous economic losses by threatening industry generation targets and long-term viability (Sarowar et al., 2013; Van West and Beakes, 2014; Iberahim et al., 2018; Sarowar et al., 2019). The World Organization for Animal Health (OIE) listed that this disease has been responsible for large-scale mortalities of farmed and wild fish in more than 20 countries across four continents (Pradhan et al., 2008; Iberahim et al., 2018). In Egypt, Aphanomyces invadans was isolated from naturally infected Striped and thin lip grey mullet (Shaheen et al., 1999), and African catfish “Clarias gariepinus” (Amany et al., 2004).
Aspergillus sp. causes systematic diseases with high death rates in fish, whereby the infections mostly occur through contamination of fish feed (Urquhart et al., 2016) and the pathogenesis of Aspergillus fumigatus and Aspergillus niger had been reported in fresh water fishes by Chauhan (2013). Fusarium is additionally viewed as imperative fish pathogens influencing marine fish and shellfish (Hatai, 2012).
The current study was conducted to detect the predominant mycotic infections with special focus on isolation, molecular identification and antimicrobial sensitivity testing for most dominant isolated fungi from cultured freshwater fishes (O. niloticus and C. gariepinus) in Kafrelsheikh fish farms.
Materials and Methods
Materials
Fish
A total number of 300 examined cultured freshwater fishes; 200 O. niloticus (120 apparently healthy and 80 showing clinical signs) and 100 C. gariepinus (30 apparently healthy and 70 showing clinical signs), were collected alive from five freshwater fish farms at Kafr El Sheikh Governorate all over the year 2018. The samples were collected with an average body weight of (40±5 and 150±10 gm) for O. niloticus and C. gariepinus, respectively.
The alive collected fishes were transferred to the lab., held in well-prepared glass aquaria supplied with sufficient amounts of dechlorinated water with continuous air supply (Innes, 1966; Langdon and Jones, 2002).
Methods
Clinical and postmortem examination
The gathered fish were clinically inspected to identify any external changes or clinical abnormalities (McVicar, 1982). Postmortem examination of the internal organs was carried out on sacrificed and freshly dead fish according to Austin and Austin (2012).
Mycological examination
Isolation of the fungus from diseased fishes
Using sterile dissecting needle, samples were taken from the skin, gills and internal organs (liver, kidney) of the collected fishes. Mycological examination was conducted according to (Noga, 1993). Gathered specimens were inoculated into Sabouraud’s dextrose broth tubes (SDB) and subsequently, inoculated into duplicate plates of Sabaroud’s dextrose agar (SDA) media with addition of chloramphenicol (50 mg/mL) to the media after autoclaving; to inhibit bacterial growth. The inoculated plates were incubated at 25±2°C for 4-7 days (Whitman, 2004). Aphanomyces sp. colonies were re-isolated on Glucose peptone yeast extract agar medium (GPYA) (Willoughby and Robberts, 1994). Negative plates were not disposed before 2 weeks (Feingold and Baron, 1986). All the positive moulds cultures examined for gross and micro morphological characteristics (Collins and Lyne, 1984; Refai, 1987).
Identification of different moulds
Studying the morphological characters of colonies included growth appearance of the cultures, rate of growth, texture and color of the surface and reverse side colonies according to Al-Dorry (1980) and Refai (1987). Preliminary recognization of moulds was conducted utilizing wet mount preparation of fish samples. A small portion of fungal colony was placed on a glass slide with drop of distilled water, the mycelial mass was gently teased a part with two dissecting needles, covered with a clean cover slide and examined under the microscope.
The confirmatory identification was carried out using cellophane tape technique. A little bit portion from the periphery of a fresh colony (3-5 days) was picked up utilizing the sticking surface of a piece of cellophane tape and placed with its stick surface down on a clean glass slide with a drop of lactophenol cotton blue stain and inspected microscopically to detect the septation of hyphe, roughness or smoothness of conidiophores, shape of vesciles, arrangement and number of rows of strigmata (Diab, 2006). Micro-slide culture technique was done on those isolates whose identification was uncertain after staining with lactophenol cotton blue (Dugan, 2006).
Molecular identification for moulds
Genomic DNA was extracted from a pure culture of fungal isolates utilizing the DNA extraction Kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. Fungal colonies were inoculated into GY broth (in case of Aphanomycess sp.) and/or SD broth (in case of Asperigillus flavus and Aspergillus niger) and incubated at 25°C for 4 days; then taken 55 mg from cultured mycelium centrifuged at 300xg for 5 minutes. Mycelia were washed with phosphate buffered saline (PBS). 20 μl proteinase K, buffer AL vortex and ethanol (96-100%) vortex were added and centrifuged at > 6000 xg (8000 rpm) for 1 min. Buffer AW1 was added and centrifuged at > 6000 xg (8000 rpm) for 1 min and buffer AW2 was added and centrifuged for 3 min at 20,000xg (14,000 rpm). The spin column was transferred to a new 2 ml micro centrifuge tube. DNA was eluted by adding 200 μl buffer AE to the center of the spin column membrane, incubated for 1 min at room temperature (15-25 C) and centrifuged for 1 min at > 6000 xg (8000 rpm). Oligonucleotide primers named as ITS1 (5′ TCCGTAGGTGAACCTGCGG 3′) and ITS4 (5′ TCCTCCGCTTATTGATATGC 3′) were designed according to White et al. (1990) for amplification of the ITS.
To amplify the DNA, a PCR reaction was prepared by adding 6 µL of extracted DNA, 1 µL of forward and reverse primers (20 pmol), 12.5 µL Emerald Amp GT PCR master mix (2x premix) and sterile water up to a final volume of 25 µL. The PCR reaction was carried out in thermal cycler (Eppendorf, Hauppauge, New York) and included an initial cycle at 94°C for 5 min, followed by 35 cycles at 94°C for 30 sec., 56°C for 40 sec., 72°C for 50 sec., and a final elongation step of 72°C for 10 min. Subsequently, PCR amplicons were separated and analyzed by gel electrophoresis by loading products onto a 1.5 % agarose gel supplemented with 1% ethidium bromide (50 ng /mL) and visualizing bands using an ultraviolet lamp at a wavelength of 300 nm.
Antifungal Sensitivity testing for isolated moulds
Antifungal sensitivity test was conducted using Mueller-Hinton agar media supplemented with 2% glucose, providing a suitable growth for most yeasts, and 0.5 mg/L methylene blue dye medium (enhances the zone edge definition) limiting the trailing impact.
The pH of the medium should be in the range of 7.2 and 7.4 after gelling and the agar should be 4 cm high. The inoculum is standardized to 0.5 McFarland using a densitometer and plates should be incubated at 35 ⁰C for 24 h; some strains show inadequate growth and may need 48 h of incubation. Two main groups of antifungals are used in the clinical setting to treat fungal infections: polyenes represented by (Amphotericin B, Nystatin,) and azoles with several derivatives such as itraconazole, fluconazole, voriconazole, clotrimazole, fluconazole (Humid).
Results and Discussion
Mycotic diseases of fish, caused by oomycete pathogens, are a real threat to the sustainability of a developing aquaculture industry. The increasing frequency of invasive fungal infection and the high mortality associated with disseminated fungal disease have highlighted the need for rapid identification of infectious molds from clinical samples.
Clinical and postmortem examination
The external gross lesions of the examined O. niloticus revealed scale detachment, fins erosion, pale body coloration and eye opacity particularly in fish naturally infected with Aphanomycess, Aspergillus, Pencillium and Fusarium. Scale detachment and fins erosion may be due to the lytic activity of primary bacterial infection as all fungal infections are considered as secondary invader pathogen; these results concurred with many authors (Refai, 1987; Bruno, 1994; Noor El-Deen et al., 2018). Fin erosion and eye opacity were mainly observed in diseased fish infected with Aphanomyces sp. where finally the fish die due to inability of the fish to feed as a result of eye opacity; the result agree with (Khalil, 1993; El-gamal, 2000; Marzouk et al., 2003; Diab, 2006; El-Atta, 2008; Bassiouny et al., 2019).
However, the infected C. gariepinus showed scattered hemorrhagic patches on the ventral abdomen; this may be credited to toxins discharged by moulds causing extreme symptomatic changes in the form of haemorrhagic patches, the result agreed with (Diab, 2006; Bassiouny et al., 2019). On the other hand, the main characteristic postmortem lesions were liver enlargement with moderate petechial hemorrhage.
Mycological examination
Mycological examination revealed the isolation of 900 fungal isolates from 120, 70 fishes showing clinical signs and 80, 30 apparently healthy O. niloticus and C. gariepinus, respectively. As shown in Table 1 and Table 4, 570 fungal isolates were isolated from O. niloticus and 330 isolates from C. gariepinus (from skin, gills, liver and kidney) with incidence percent of (63.3% and 36.7%) among O. niloticus and C. gariepinus, respectively. The high recurrence of mould isolates in O. niloticus concurs with Diab (2006), Refai et al. (2010) and Bassiouny et al. (2019).
Table 1: Total prevalence of moulds infections among O. niloticus and C. gariepinus
| Fish species |
No. of examined fish |
No. of diseased fish | No. of apparently healthy fish | No. of mould isolates | percentage |
| O. niloticus | 200 | 120 | 80 | 570 | 63.3% |
| C. gariepinus | 100 | 70 | 30 | 330 | 36.7% |
| Total | 300 | 190 | 110 | 900 | 100% |
Morphological identification of isolated moulds
The morphological identification of moulds revealed the presence of different moulds including: Aphanomyces sp., Aspergillus flavus; Aspergillus niger; Penicillium sp.; Fusarium sp., Mucor sp. and Alternaria sp. The most prevalent isolated fungi was Aphanomyces sp. (162 isolate representing 28.4%) (100 isolates representing 30.3%) mainly from skin, fins and gills from O. niloticus and C. gariepinus, respectively showing the highest incidence among diseased fishes. However, the most prevalence isolated fungi from apparently healthy fishes were Aspergillus flavus (55, 34), Aspergillus niger (60, 26), Mucor sp. (29, 25) and Alternaria sp. (28, 10) mainly from internal organs of O. niloticus and C. gariepinus, respectively; the results concur with previously investigated data (Fafioye et al., 2008; Refai et al., 2010; Bassiouny et al., 2019). Refai et al. (2010) has characterized Aspergillus sp., Penicillium sp. and Rhizopus sp. as normal mycoflora and these species may be considered as opportunistic pathogens as many of these genera possess virulence factors which enable them to cause disease especially under favorable conditions.
As shown in (Tables 2A, 2B), mycological examination demonstrated the incidence of Aphanomyces sp. (143, 19), Aspergillus flavus (26, 55), Aspergillus niger (19, 60), Penicillium sp. (24, 28), Fusarium sp. (18, 11) Mucor (50, 29) and Alternaria sp. (60, 28) in diseased and apparently healthy O. niloticus, respectively. On the other side, mycological examination of C. gariepinus demonstrated the incidence of Aphanomyces sp. (80, 20), Aspergillus flavus (15, 34), Aspergillus niger (25, 26), Penicillium sp. (10, 20), Fusarium sp. (7, 3) Mucor (15, 25) and Alternaria sp. (40, 10) in diseased and apparently healthy C. gariepinus, respectively (Table 3A, 3B).
Table 2A: Incidence of moulds isolated from different organs of diseased O. niloticus.
| Moulds | O. niloticus | ||||
| Skin & fins | Gills | Liver | Kidney | ||
| No. | No. | No. | No. | No. | |
|
Aphanomyces sp. |
143 | 109 | 34 | 0 | 0 |
| Asperigillus flavus | 26 | 0 | 0 | 26 | 0 |
| Asperigillus niger | 19 | 0 | 0 | 15 | 4 |
|
Pencillium sp. |
24 | 12 | 8 | 4 | 0 |
|
Fusarium sp. |
18 | 0 | 18 | 0 | 0 |
|
Mucor sp. |
50 | 25 | 17 | 6 | 2 |
|
Alternaria sp.. |
60 | 35 | 21 | 4 | 0 |
| Total | 340 | 181 | 98 | 55 | 6 |
Table 2B: Incidence of moulds isolated within different organs of apparently healthy O. niloticus.
| Moulds | O. niloticus | ||||
| Skin & fins | Gills | Liver | Kidney | ||
| No. | No. | No. | No. | No. | |
|
Aphanomyces sp. |
19 | 11 | 8 | 0 | 0 |
| Asperigillus flavus | 55 | 0 | 0 | 50 | 5 |
| Asperigillus niger | 60 | 8 | 11 | 13 | 28 |
|
Pencillium sp. |
28 | 12 | 8 | 5 | 3 |
|
Fusarium sp. |
11 | 0 | 11 | 0 | 0 |
|
Mucor sp. |
29 | 13 | 13 | 3 | 0 |
|
Alternaria sp. |
28 | 10 | 8 | 6 | 4 |
| Total | 230 | 54 | 59 | 77 | 40 |
The colonies of Aphanomyces sp. appeared white and transparent with velvet surface (become heavy and opaque at 7th day); while, microscopically by wet mount appeared as branched non-septated hyphae and when stained with lactophenol cotton blue, gave non septated, y- shape branched hyphae with cytoplasmic organelles (Figure 1). The result concurs with that reported by Abd El-Latif (2003), Afzali et al. (2013) and Youssuf et al. (2017).
Table 3A: Incidence of moulds isolated from different organs of diseased C. gariepinus.
| Moulds |
C. gariepinus |
||||
| Skin & fins | Gills | Liver | Kidney | ||
| No. | No. | No. | No. | No. | |
| Aphanomyces invadans | 80 | 55 | 25 | 0 | 0 |
| Asperigillus flavus | 15 | 0 | 0 | 15 | 0 |
| Asperigillus niger | 25 | 5 | 3 | 10 | 7 |
|
Pencillium sp. |
10 | 5 | 2 | 3 | 0 |
|
Fusarium sp. |
7 | 0 | 7 | 0 | 0 |
|
Mucor sp. |
15 | 10 | 5 | 0 | 0 |
|
Alternaria sp. |
40 | 8 | 13 | 13 | 6 |
| Total | 192 | 83 | 55 | 41 | 13 |
Table 3B: Incidence of moulds isolated from different organs of apparently healthy C. gariepinus.
| Moulds | C. gariepinus | ||||
| Skin & fins | Gills | Liver | Kidney | ||
| No. | No. | No. | No. | No. | |
| Aphanomyces invadans | 20 | 13 | 7 | 0 | 0 |
| Asperigillus flavus | 34 | 0 | 0 | 30 | 4 |
| Asperigillus niger | 26 | 4 | 4 | 6 | 12 |
|
Pencillium sp. |
20 | 10 | 7 | 3 | 0 |
| Fusarium sp. | 3 | 0 | 3 | 0 | 0 |
| Mucor sp. | 25 | 10 | 7 | 8 | 0 |
| Alternaria sp. | 10 | 2 | 4 | 4 | 0 |
| Total | 138 | 39 | 32 | 51 | 16 |
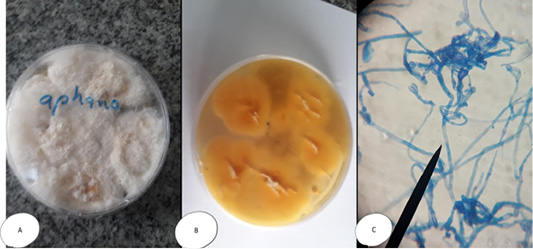
Figure 1: Aphanomyces invadans (A) Petri dish Surface side (B) Petri dish reverse side - with the characteristic white transparent velvet surface (become heavy and opaque at 7th day on SDA). (C) hyphae stained with LCB give non septated branched hyphae with cytoplasmic organelles, the branches of hyphae give y- shape.
Interestingly, Aspergillus species exhibited some variation within the same genus. Aspergillus flavus (A. flavus) appeared velvety with numerous aerial growths; the color changes from yellow to yellowish green by aging; while microscopically, the conidiophores were long and rough, the vesicles were rounded and large and the strigmata were biseriate, loose, and radiate giving rise to ovoid rough conidia (Figure 2). Colonies of Aspergillus niger (A. niger) had black color with radiated edges with wooly texture, while microscopically had extremely long, smooth and yellowish conidiophores, vesicles were very large and globes while the strigmata were biseriate, compact and radiate and the conidia were globes and smooth (Figure 3).
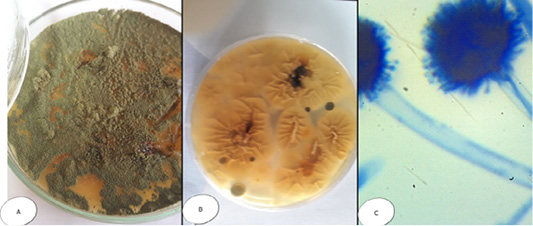
Figure 2: Aspergillus flavus (A) Petri dish…Surface side (B) Petri dish reverse side (C) hyphae stained with LCB showing characteristic typical head.
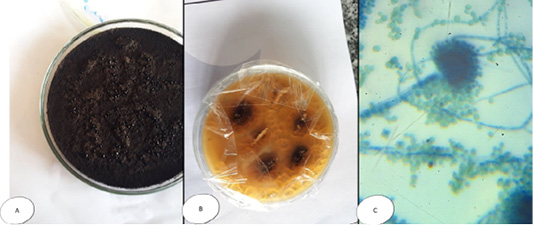
Figure 3: Aspergillus niger (A) Petri dish Surface side (B) Petri dish….reverse side (C) hyphae stained with LCB showing characteristic round head with black conidia.
Pencillium sp. colonies white and fleecy transformed into greenish blue in colour, while microscopically, septated hyphae with un branched condiophores possessing metula with flask-shaped strigmata and the metulae bear un branched chain of conidia; the entire structure form brush appearance (Figure 4). Regardless Fusarium sp., colonies were cottony or wooly in texture, snow white, pink-violet or rosy-red in color, with scattering of colored pigments into the reverse surface of the medium; while microscopically, they appeared long, branched and septated hyphae from which short conidiophores rose singly or in groups, and sometimes branched. Two types of conidia were observed, a large banana-shaped, septated macroconidia and small, round, non septated microconidia (Figure 5).
Colonies of Mucor sp. appeared fast-growing, white-to-gray cotton candy, became dark with time and fills the petri dish with fluffy mycelium; and microscopically, non-septated wide hyphae. Sporangiophores are long, may be branched and end with bear terminal round sporangia. The sporangia have a thin wall which when mature dissolves (or is disrupted) to discharge round or somewhat ellipsoidal sporangiospores (4-8µm diameter). With the spores dispersed. No rhizoids are formed (Figure 6).
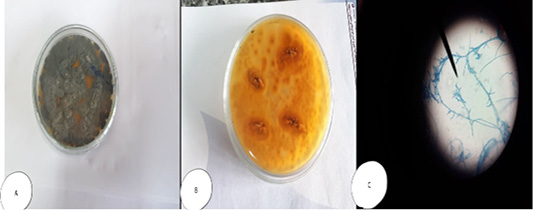
Figure 4: Penicillium sp. (A and B) Petri dish on SDA with different colour and texture. (C) hyphae stained with LCB showing characteristic brush- like arrangement.
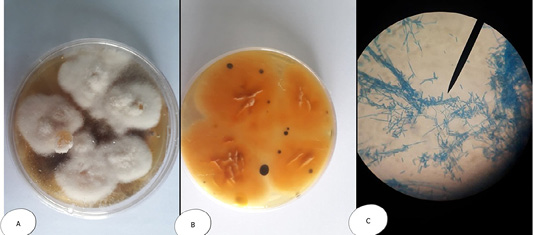
Figure 5: Fusarium sp. (A) Petri dish Surface side (B) Petri dish reverse side (C) hyphae stained with LCB showing characteristic snow white colonies with rose pigment in the center.

Figure 6: Mucor sp. (A) Petri dish…Surface side (B) Petri dish reverse side (C) hyphae stained with LCB showing characteristic round sporangia.
Alternaria sp. revealed grayish brown to green color colonies; while microscopically appeared as septated hyphae with dark branched condiophores creating at least two expanding tree-like with oval conidia having velvety dull gray colour (Figure 7). The observed colonies of all isolated moulds, in the current study, are in accordance with previous reports (El-gamal, 2000; Ali, 2007; Refai et al., 2010; Bassiouny et al., 2019).
Molecular identification
Molecular identification (Polymerase Chain Reaction) using inter-transcriped spacer (ITS) gene, which is a universal gene,
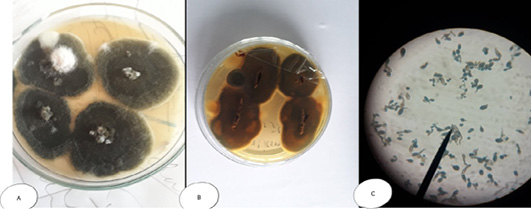
Figure 7: Alternaria sp. (A) SDA Petri dish Surface side showing grayish brown to green color (B) Petri dish reverse side (C) hyphae stained with LCB showing characteristic septated hyphae with dark branched condiophores producing two or more branching tree-like with oval condia having velvety dull gray colour under microscope.
is one of the most important approaches to distinguish between different fungal infections (Eissa et al., 2013). Therefore, in the current study, molecular identification was performed using inter-transcriped spacer (ITS) gene for the most prevalent isolates among either diseased or apparently healthy fishes. Three Aphanomyces invadans isolates from diseased C. gariepinus and two isolate of Asperigillus flavus and Asperigillus niger isolates from apparently healthy O. niloticus were used. Developed PCR assay specific primer detect not only Aphanomyces invadans showing clear bands at 569 bp molecular weight (Figure 8), but also Asperigillus flavus and Asperigillus niger showing clear bands at 595 and 600 bp molecular weight, respectively (Figure 9). The current obtained result is nearly similar to Oidtmann et al. (2008) and Youssef (2017) where Aphanomyces invadans produced a clear band at 564 bp and 600 bp, respectively. On the other hand, Afzali et al. (2015) reported that the result of PCR assay of Aphanomyces invadans showed clear band at 400 bp. These variations may be attributed to various tested primers sequences and PCR protocol as a result of the different amplification cycles (Oidtmann et al., 2008; Afzali et al., 2015; Youssef, 2017). On the other hand, the result of PCR assay of Asperigillus flavus and Asperigillus niger was exactly similar to Henry et al. (2000) where Asperigillus flavus and Asperigillus niger produced a clear band at 595 bp and 599 bp, respectively.
Antifungal sensitivity test
Antifungal susceptibility testing methods was applied to distinguish antifungal resistance and to decide the best treatment for a particular fungus. Clinical microbiology relies on these techniques to select the agent of choice for a fungal infection, and to know the local and the global epidemiology of antifungal sensitivity.
Anti-fungal sensitivity test was conducted using the disc diffusion method for the isolated strains. As demonstrated in Table 5, all mould isolates are sensitive to Nystatin and the majority of them were resistant to Voriconazole. The results agree with Ebaied (2014) where it was investigated that Asperigillus flavus, Asperigillus niger, Penicillium and Alternaria were sensitive to Nystatin.
Table 4: Incidence of Moulds isolated from O. niloticus and C. gariepinus.
| Isolates | O. niloticus | C. gariepinus | Total isolates No. | % in relation total isolate No. | ||
| No. | % | No. | % | No | % | |
| Aphanomyces invadans | 162 | 28.4 % | 100 | 30.3% | 262 | 29.1% |
| Asperigillus flavus | 81 | 14.2 % | 49 | 14.8 % | 130 | 14.4 % |
| Asperigillus niger | 79 | 13.9% | 51 | 15.5 % | 130 | 14.4 % |
| Pencillium sp. | 52 | 9.1% | 30 | 9.1% | 82 | 9.1% |
| Fusarium sp. | 29 | 5.1% | 10 | 3.0% | 39 | 4.4 % |
| Mucor sp. | 79 | 13.9 % | 40 | 12.1% | 119 | 13.2 % |
|
Alternaria sp. |
88 | 15.4 % | 50 | 15.2% | 138 | 15.4 % |
| Total Moulds | 570 | 100% | 330 | 100% | 900 | 100% |
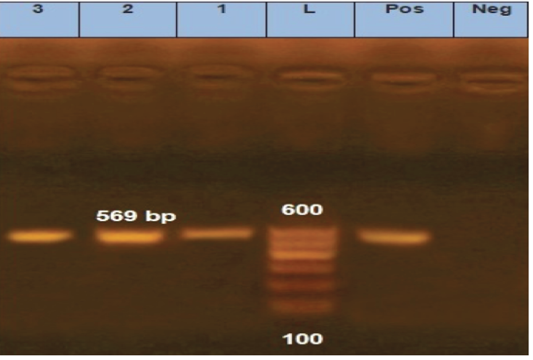
Figure 8: Agarose gel electrophoresis for PCR products representing amplification of 569 bp of inter-transcribed spacer (ITS) gene in Aphanomyces invadans. Lane 1: 100 bp DNA Ladder. Pos.: is control positive; Neg.: is control Negative; Lane 1, 2 and 3: positive sample.
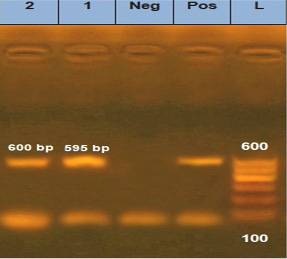
Figure 9: Agarose gel electrophoresis for PCR products representing amplification of 595 bp of inter-transcribed spacer (ITS) gene in Asperigillus flavus and 600 bp Asperigillus niger. Lane 1: 100 bp DNA Ladder; Pos.: is control positive; Neg.: is control Negative; Lane 1: positive sample for Asperigillus flavus; Lane 2: positive sample for Asperigillus niger.
Table 5: Results of Antifungal susceptibility test of Moulds isolated from fresh water fish.
| Mould species |
NS ( 10 ) |
AP (100) |
CC ( 10 ) |
IT (10 ) |
FLC ( 10 ) |
VRC (10 ) |
| Aphanomyces sp. | S | S | S | S | R | R |
| Asperigillus flavus | S | R | S | S | R | R |
| Asperigillus niger | S | S | R | S | R | S |
| Pencillium sp. | S | R | R | R | R | R |
| Fusarium sp. | S | S | S | S | S | R |
| Mucor sp. | S | S | S | S | S | R |
| Alternaria sp. | S | S | S | S | S | S |
NS: Nystatin (10mcg/disc) IT: Itraconazole (10mcg/disc); AP: Amphotericin B (100mcg/disc) FLU: Fluconazole (10mcg/disc); CC: Clotrimazole (10mcg/disc) VRC: Voriconazole (10mcg/disc); S: sensitive R: resistant.
Conclusion
From the current study, it could be concluded that Aphanomyces invadans was the predominant fungi among diseased fishes mainly from skin, fins and gills. However, Aspergillus flavus, Aspergillus niger and Alternaria sp. were the most predominant fungi isolated from apparently healthy Oreochromis niloticus and Clarias gariepinus mainly from internal organs. In brief, although most fungi are considered to be normal mycoflora, but they still can cause infections to fishes. Therefore, it is suggested that proper health management practices should be adapted while rearing fish, so that chances of fungal infection can be minimized.
Authors Contribution
All authors contributed equally.
Conflict of interest
The authors declare no conflicts of interest.
References






