Advances in Animal and Veterinary Sciences
Research Article
Genetic Characterization of Mycoplasma. bovis, L. moncytogenes and Brucella species Recovered from Bovine Abortion
Walid S. Mousa1*, Yousreya M. Hashem2, Hamed T. Elbaz3, Eman E. Abdeen4, Sherifa Mostafa M. Sabra5; Eman I. Beleta6, Mohamed A. Nayel1
1Department of Animal Medicine and Infectious Diseases, Faculty of Veterinary Medicine, University of Sadat City, 32897, Egypt; 2Mycoplasma Department, Animal Health Research Institute, Agricultural center, Dokki, Giza, Egypt, 12618; 3Department of Theriogenology, Faculty of Veterinary Medicine, University of Sadat City, 32897, Egypt; 4Department of Bacteriology, Mycology and Immunology, Faculty of Veterinary Medicine, University of Sadat City, 32897, Egypt; 5Serology Unit, Animal Health Research Institute, Agricultural Research Center, Dokki, Giza, 12618 Egypt; 6Brucella Department, Animal Health Research Institute, Agricultural Research Center, Dokki, 12618, Egypt.
Abstract | Abortion is one of the devastating and economic problems in dairy sector. This study aimed to determine the common bacterial cause of bovine abortion in Menoufiya governorate, Egypt from October 2018 till December 2019. Out of examination of 600 cows, 100 aborted cases (16.7%) were observed during this period and 55 vaginal swabs, 15 fetal stomach contents and 30 milk samples were collected from aborted cows and subculture onto specific medium. Our findings concluded that bacterial isolation reported that B. melitensis and L. monocytogenes were the most isolated from 55 vaginal swabs with 3.6% followed by M. bovis 1.8%. Meanwhile B. abortus and B. melitensis were the most common with 13.3% from 15 foetal stomach content followed by L. monocytogenes 6.6%. Moreover, M. bovis and B. abortus, and L. monocytogenes were isolated from 30 milk samples with 6.6%. The PCR results revealed successful amplification of B. abortus, B. melitensis, M. bovis and L. monocytogenes at 498, 731, 360, and 938 respectively. The sequencing analysis of B. melitensis, B. abortus, L. monocytogenes and M. bovis demonstrated identical and strong similarities (96.67-100%) with many international field strains in the Gene bank. To the best of our knowledge, this was the first report for L. monocytogenes and M. bovis from aborted fetus that showed high homologues with many international field stains. In conclusion, B. melitensis, B. abortus, M. bovis and L. monocytogenes, were the most common bacterial causes of bovine abortion in Egypt, thus promoting more attention and studies to establish new methods for control measures.
Keywords | Abortion, Brucella, L. monocytogenes, M. bovis, Sequencing.
Received | March 12, 2021; Accepted | March 17, 2021; Published | June 15, 2021
*Correspondence | Walid S Mousa, Department of Animal Medicine and Infectious Diseases, Faculty of Veterinary Medicine, University of Sadat City, 32897, Egypt; Email: walidsaadvet@yahoo.com
Citation | Mousa WS, Hashem YM, Elbaz HT, Abdeen EE, Sabra SMM, Beleta EI, Nayel MA (2021).Genetic characterization of mycoplasma. bovis, l. moncytogenes and brucella species recovered from bovine abortion Adv. Anim. Vet. Sci. 9(7): 1012-1019.
DOI | http://dx.doi.org/10.17582/journal.aavs/2021/9.7.1012.1019
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2021 Mousa et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Abortion is the most serious and economical issue disturbs the reproductive efficacy leading to a decrease in milk production, replacement of herd, expensive care and a rise in culling rate (Tulu et al., 2018). Several bacterial agents such as brucella, listeria, leptospira species, and viral agents such as BVD, IBR as well as mycotic and protozoal agents are the major infectious causes of bovine abortion. In addition, genetic, management, environmental and geographical factors are also included in non-infectious determinants (Tulu et al., 2018). Abortion typically takes place between 42 and 260 days of pregnancy (Peter, 2000). More than 2-5% of abortion rate is considered a serious herd issue that often varies greatly depending on the production nature, animal breed and state of the husbandry management. Therefore, more attention was needed to establish the definitive etiology and preventive control strategy (Eshete and Moges, 2014). Brucellosis is a common serious zoonotic disease among various animal species and humans resulted in abortion, retained placenta, premature birth and infertility disorders (Muma et al., 2007). B. abortus, B. melitensis and B. suis are the common strains that continue to spread the infection among dairy herds (Aznar et al., 2014). The pathogenicity of Brucella relies primarily on interference with the immune system by adapting oxygen limited to macrophages (Lopez-Goni et al., 2002). As well as many associated virulence genes such as omp31 and omp31 genes are also included in Brucella pathogenicity mechanism (Ramadan et al., 2019). In parallel to serological studies, arrange of molecular techniques are essentially important for the design of epidemiological maps and phylogenetic homogeneity of circulating clones and successful control programs (El-Diasty et al., 2018). L. monocytogenes is the primary cause of listeriosis in ruminants characterized by three distinct forms; septicemia, encephalitis and abortion as well as mastitis, repeat breeding and endometritis (Malik et al., 2002). Mycoplasma bovis is fastidious organism that causes many disease syndromes including pneumonia, mastitis, arthritis, otitis media, and reproductive disorders (Maunsell et al., 2011). Additionally, (Ghanem et al., 2013) described that M. bovis had emerged role in reproductive and infertility disorders such as vulvovaginitis and endometritis. However, (Abdeen et al., 2017) declared that M. bovis is a major infectious agent involved in respiratory diseases in bovine animals and the sequencing of M. bovis is more powerful method for diagnosis and mapping foreign linages of the circulating strains. The traditional diagnosis of Mycoplasma bovis is a confirmatory method but time consuming and cross reactivity is a common problem (Kumar et al., 2001). The significance and economic importance of bovine abortion in cattle sector industry encourage continuous researches and interest. Therefore, this current study was focused to identify the major bacterial species causing bovine abortion in Egypt. In addition to sequencing and phylogenetic analysis of the obtained strains with the local or foreign clones.
Material and methods
Samples collection
Examination of six-hundred cows of mixed breed cattle from the period October 2018 till December 2019 around Sadat City area from small scales breeders. The examined cows are reared in small-scales in an individual and separate areas and the applied hygienic practice is low as well as the vaccination programs against infectious diseases is not strictly controlled. One-hundred cows were aborted after the 5th month of pregnancy till parturition time during this period. Various types of samples, vaginal discharge (55); foetal stomach content (15), and milk (30) were obtained and collected from aborted cows under aseptic condition in cool icebox and transmitted to the laboratory as soon as possible for further examination.
Isolation and Phonotypic identification of Brucella, Mycoplasma and Listeria species
All samples types (Vaginal discharge, foetal stomach content and milk) were processed and cultivated onto selective Brucella medium base (Oxiod, Code CM0169) with 5-10% v/v inactivated horse serum (SR 0035) and 1-5% of a sterile glucose solution. All cultured samples were incubated at 37 °C for successive 5-7 days with and without 5-10 % CO2 as described by (Alton et al., 1988). The phenotypic morphology and biochemical tests for genotyping of the Brucella species were done according to (MacMillan, 1990). At the same time, all samples were immersed in 2 ml of Mycoplasma broth medium (Thermo Fisher Scientific Inc) at 37°C and then plated onto specific PPLO medium (Thermo Fisher Scientific Inc) with 5% CO2 for 3 days. The microscopic minute visible transparent colonies of fried egg colonies were subjected to biochemical confirmation of Mycoplsama and serotyping was performed as described by (Nicholas and Ayling, 2003). For the isolation of Listeria species, Buffered Listeria Enrichment Broth (BLEB) was used as pre-enrichment at 30°C, for 48 hours, followed by specific plating on Oxford medium and Sheep blood agar up to 48 hours at 35 °C as described by (Scotter et al., 2001). Typical identification and standard biochemical activities of phenotypic colonies were performed as described by (Aygun and Pehlivanlar, 2007). Other common bacterial causes of abortion (Leptospira spp, Salmonella spp, Campylobacter spp) were omitted through negative culture onto specific medium for each organism.
Molecular detection of B. abortus, B. meltensis, L. monocytogenes and M. bovis
DNA extraction: DNA was extracted using commercial kits (QIAamp DNA mini kit; Qiagen, Hilden, Germany) according to manufacturer recommendations. PCR assay conditions were done as the following: The reaction mixture consisted of, PCR Master Mix (Thermo Scientific) (2X solution consisted of Taq DNA Polymerase, dNTPs, mgcl2) 0.2 μM of each primer (Metabion international AG, Germany) and complete by DNA RNAase free water, the reaction mixture was dispensed into Micro-Amp vials (47 μl per tube). The samples were cycled in a thermocycler (S-1000 BIO-RAD PCR thermocycler). The products (5μl from each reaction mixture) were analyzed by electrophoresis through a 1.5% stained with ethidium bromide agarose gel, after which the gel was photographed.
Phylogenic and sequencing analysis for B. abortus, B. melitensis, L. monocytogenes and M. bovis
The amplified fragments were purified using Gene Jet PCR purification kit; thermo scientific (Cat no. KO701). Sequencing of the PCR product on GATC Company by use ABI 3730xl DNA sequencer by using forward and reverse primers. The identification of homologies between nucleotide and amino acid sequences of our strains of accession numbers (BMMen1, BAMen1, LMEG1, and MBab1) for B. meltensis, B. abortus, L. monocytogenes, and M. bovis respectively were compared with other strains published on GenBank using BLAST 2.0 and PSI- BLAST search programs, (National Center for Biotechnology Information NCBI (http://www.ncbi.nlm.nih.gov/) respectively. The obtained nucleotide sequences comparisons and their multiple alignments with reference strains were done.
Results
Prevalence of B. abortus, B. melitensis, L. monocytogenes and M. bovis recovered from aborted cattle cases
Out of examination of six-hundred cows; 100 (16.7%) cows were aborted after the fifth month of pregnancy. Bacteriological examination of 55 vaginal discharge reported that B. melitensis and L. monocytogenes were the most predominant bacteria with 3.6% for each while M. bovis 1.8%. In contrast, B. abortus and B. melitensis were the most identified bacterial species with prevalence rate 13.3% from 15 foetal stomach content followed by L. monocytogenes 6.6%. Moreover, M. bovis and B. abortus, and L. monocytogenes were isolated from 30 milk samples with 6.6% for each as shown in Table (2).
Molecular detection of B. abortus, B. melitensis, L. monocytogenes and M. bovis recovered from aborted cattle
The molecular detection of B. abortus, B. melitensis, L. monocytogenes and M. bovis was successfully amplified theses strains by using of specific primer sets at 498,731, 938, and 360 bp as shown in Fig (1, 2, 3, & 4) respectively.
Sequencing and Phylogenic analysis of B. melitensis, B. abortus, L. monocytogenes and M. bovis recovered from aborted cattle cases
Sequencing and Phylogenic analysis of B. melitensis recovered from aborted cattle cases:
In our study, the phylogenetic analysis of our B. melitensis strain described identical genetic similarities with a number of foreign strains from China and India (100%). The strain (VB12455) of human blood sample in India was extremely similar with our strain (BMMen1) and also with
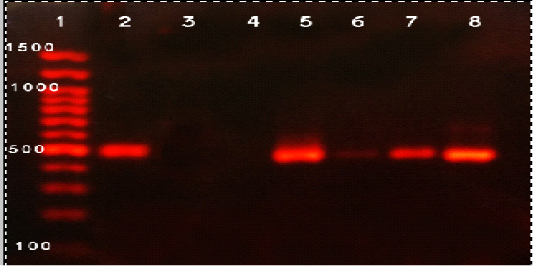
Figure 1: Agarose gel electrophoresis of PCR products of B. abortus isolates. Lane 1: 100 bp DNA, lane 2: control positive, lane 3: control negative, lane 4-8 positive B.abortus samples
ladder; lanes 2, Positive control, Lane 3 Negative control, lane 4: Negative sample, Lanes 5-8: positive samples.
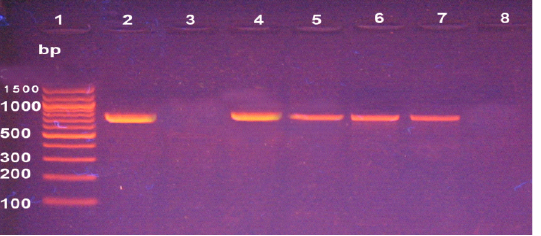
Figure 2: Agarose gel electrophoresis of PCR products of B. Melitensis isolates. Lane 1: 100 bp DNA ladder; lanes 2, Positive control, Lane 3 Negative control, lanes 4-7: positive samples, Lane 8: Negative sample.
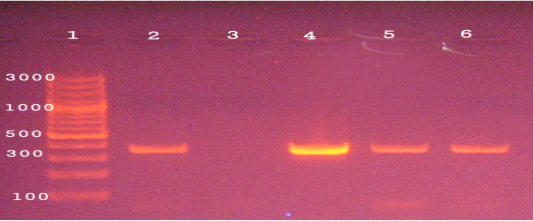
Figure 3: Agarose gel electrophoresis of PCR products of 16S rRNA gene of M. bovis (360 bp). Lane 1: 1000 bp ladder, Lane 2 control positive, Lane3 control negative, Lane 4-6 positive to M. bovis.
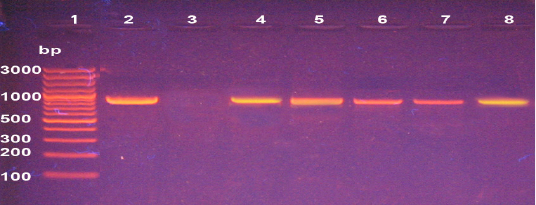
Figure 4: Agarose gel electrophoresis of PCR products of 16S rRNA gene of L. monocytogenes (938 bp). Lane 1: 1000 bp ladder, Lane 2 control positive, Lane 3 control negative, Lane 4-8 positive to Listeria.
2 strains (VB700 and C11MS-NV-1) from human blood and animal origin specimen from India. In addition, two strains from China showed identical of homologues with our strain on Gen bank (Xy-Bru and QH61) from environmental source and aborted fetus of yak as shown in Fig (5).

Figure 5: Dendrogram showed the genetic homogeneity of B. melitensis strain (BMMen1) from aborted cows with other related international isolates.
Sequencing and Phylogenic analysis of B. abortus recovered from aborted cattle cases:
B. abortus strain in our study (BAMen1) found extremely genetic similarity (96.67-100%) with four different strains from different countries. As well as a strain (C11MS-NV-4) of animal origin in India. Additionally, one vaccine-origin strain from China of (104M). Finally, identical homologues and familiar similarities with the Egyptian local strain (Egy19) of the aborted cow milk origin as shown in Fig (6).
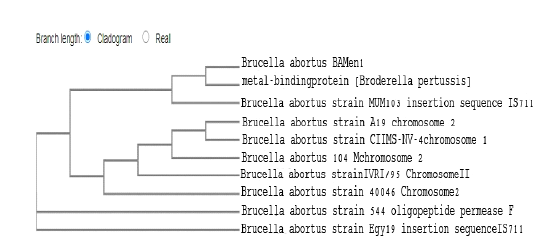
Figure 6: Dendrogram showed the genetic homogeneity of B. abortus strain (BAMen1) from aborted cows with other related international isolates.
Sequencing and Phylogenic analysis of L. monocytogenes recovered from aborted cattle cases:
In our study, our strain (LMEGY1) reported that the phylogenetic analysis of L. monocytogenes strain revealed nearly identical similarities (99.78-100%) with several foreign strains. Identical similarities were found with the strains (Ka89-2) isolated from raw minced meat in Turkey and the strain (G1KNSWL31) from water sources in South Korea. Additionally, high identical similarities with human clinical specimens in the USA of the two strains (L. monocytogenes 52330; LAMP18-H8393) with local Egyptian strain of accession number (EB51), and a strain (NCTC7974) from human blood specimen in UK as shown in Fig (7).
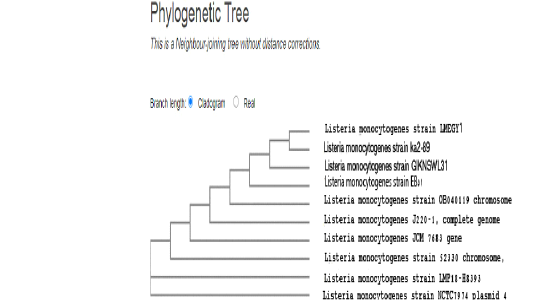
Figure 7: Dendrogram showed the genetic homogeneity of L. monocytogenes strain (LMEGY1) from aborted cows with other related international isolates.
Sequencing and Phylogenic analysis of M. bovis recovered from aborted cattle cases:
The sequencing analysis of M. bovis strain in our study (MBab1) clustered identical homologs similarity with one strain of bovine origin in Hungary. Meanwhile, high genetic links were also found with two Canadian strains from joint of cattle breed (Bos Taurus) and lung of feeder bull (Bison bison). Additionally, three strains from china, Japan, and Belgium of separate geographic areas of accession number (16M, KG4397, and VK1 16 S rRNA) respectively showed high sequencing similarities. In addition, obvious segregation of one strain (NADC61) from bull lung in Canada as shown in Fig (8).

Figure 8: Dendrogram showed the genetic homogeneity of M. bovis strain (MBab1) from aborted cows with other related international isolat
Discussion
Abortion is a significant fertility problem and a major impediment to the growth of animal production (Ararsa and Wubishet, 2014) In addition, it has a strong adverse effect on the gynecological and reproductive output of dairy herds (Ernest, 2009). In bovine abortion, multiple viruses,
Table 1: List of primer pairs and cycling conditions for the B. abortus, B. melitensis M. bovis and L. monocytogenes used in this study
| Organism | M. bovis | B. abortus & B. melitensis |
L. monocytogenes ( 16SrRNA) |
|
Primer pairs(5'-3')
|
CCT TTT AGA ITG GGA TAG CGG ATG CCG TCA AGG TAG CAT CAT TTC CTA T |
B. abortus –GACGAACGGAATTTTTCCAATCCC B. melitensis TGCCGATCACTTAAGGGCCTTCAT IS711-pecific:AAATCGCGTCCTTGCTGGTCTGA |
LI1 5..–CTC CATAAA GGT GAC CCT- d U1 5..–CAG CMG CCG CGGTAATWC |
| PCR product (bp) | 360 bp |
B. abortus (498 bp) B. melitensis (731 bp); |
938 bp |
|
Cycling conditions
|
95°C for 3mins, (35 cycles): Denaturation at 94 °C for 45 s. Annealing at 60 °C for 60 s. Polymerization at 72 °C for 2min. Final extension at 72 °C for 3 min. |
95°C for 3mins, (35 cycles): Denaturation at 95 °C for 1.15min Annealing at 55.5 °C for 2 min Polymerization at 72 °C for 2 min Final extension at 72 °C for 10 min |
95°C for 3mins, (35 cycles): Denaturation at 94°C for 30 sec. Annealing at 53 °C for 15 sec Polymerization at 72 °C for 90 sec. Final extension at 72oC for 7min |
Table 2: Prevalence of B. abortus, B. melitensis, L. monocytogenes and M. bovis recovered from aborted cattle cases
| Samples | Number of samples | M.bovis | B.abortus | B.melitensis | L. monocytogenes | ||||
| No | % | No | % | No | % | No | % | ||
| Vaginal discharge | 55 | 1 | 1.8 | 0 | 0 | 2 | 3.6 | 2 | 3.6 |
| Fetal stomach content | 15 | 0 | 0 | 2 | 13.3 | 2 | 13.3 | 1 | 6.6 |
|
Milk |
30 | 2 | 6.6 | 2 | 6.6 | 0 | 0 | 2 | 6.6 |
| Total | 100 | 3 | 4 | 4 | 5 | ||||
bacteria, protozoa and mycotic causes are included. Surveillance about the accurate rate of infectious agents is not really estimated, although infectious agent was detected in about 90% of cases (Parthiban et al., 2015). In addition, heat and production stress, seasonal, maternal and paternal are non-infectious factors that probably must put in consider (Lee and Kim, 2007). In the present study, the over prevalence rate of abortion rate was 16.7%. Nearly similar findings were reported in Uganda 14% (Miller et al., 2016), also in Ethiopia the abortion rate in cattle ranged from 2.2 to 28.9%, (Siyoum et al., 2016). On the other hand, lower prevalence was reported in Pakistan 6.3% (Ali et al., 2017). Higher prevalence rate 33.5% was recorded in Morocco by (Lucchese et al., 2016). In this study the most isolated and identified bacteria were L. monocytogenes, B. abortus, B. melitensis, as well as M. bovis with prevalence rate ranged from 3 - 5% as shown in Table (1). This was in contrary with study of (Derdour et al., 2017) who revealed that abortion rate due to B. abortus in Algeria was 3.06%. While, higher prevalence rate of B. abortus 14.2% was found in Brazil from various samples (Silva et al., 2009). In addition, (Ramadan et al., 2019) recorded that B. melitensis bv3 was most prevalent in Egypt with prevalence rate 2.33%.
Regarding to several studies described that M. bovis as a serious cause of several disease syndromes such as mastitis, arthritis, otitis media, and reproductive disorders causing significant economic losses in cattle herds (Nicholas, 2011) and the genital disorders commonly reported intermittent outbreaks (Pfutzner and Sachse, 1996). Similar results showed that L. moncytogenes in case of spontaneous abortion was 3.3% (Aygun and Pehlivanlar, 2006). In another study (Eslami et al., 2014) recorded higher prevalence rate of L. monocytogenes 7.2% and 12.5% by isolation and PCR from aborted human cases. Difference in abortion rates between the published studies may be attributed to variation in cattle breeds, sample size, production status, and environment and management factors. Moreover, other husbandry conditions in the studied area such as the small scales breeding system at which the cows are reared in small separate and low hygienic condition and the vaccination programs for control of infectious diseases is not so firmly applied or control, thus allow more spreading and dissemination of the infections between animals in neighboring areas through the animal movement and animal trading.
The application of PCR was in this study is more efficient and successfully amplified of B. abortus, B. melitensis M. bovis, and L. monocytogenes at 498, 731, 360 and 938 bp respectively as shown in Fig (1, 2, 3, & 4). Previous studies have showed that PCR is an efficient technique for the diagnosis of bovine brucellosis (Kaushik et al., 2006). In addition, (Bricker et al., 2003) declared that PCR and RT-PCR were evolved for detection of several infectious bacteria causing bovine abortion such as B. abortus and M. bovis. Furthermore, (Caswell et al., 2010) identified that most of molecular techniques are focused on the sequences of specific DNA of the uvrC and oppD/F genes particularly the uvrC gene is able can distinguish between M. bovis from other similar mycoplasma species. In a comparative study in Egypt, (El-Sayed et al., 2019) standardized different four pairs of primers to amplify the 31 KDa gene encoding protein in Brucella spp., lig gene in pathogenic Leptospira, hlyA gene in L. monocytogenes and 16S rDNA in Mycoplasma spp.
It is important to further explore the rapid development and advances in molecular techniques, such as PCR, RT- PCR, mPCR, as well as gene sequencing and phylogenetic analysis to achieve their maximum functional potential (Verma et al., 2015). Sequencing analysis in this study was carried out for a single strain from each B. melitensis and B. abortus from abortion cases and the result revealed a strong genetic similarity (96.67%- 100%) with several strains from various regional nations and localities. This outcome was consistent with the analysis of 69 Egyptian strains of B. melitensis and B. abortus strains byMLVA-16 technique with other Mediterranean Sea strains (Wareth et al., 2020) who found that B. melitensis strains were clustered into two main cluster containing 21 genotypes, with three major clusters include nine genotypes for B. abortus strains. Also, the authors reported that the irregularity in distribution of these strains over time and areas in the Mediterranean Sea countries. Interestingly, 16 patterns of B. melitensis from Egypt were found in closely related to Italian strains as well as B. abortus strains shared with the same genotype with Portugal and Italy strains. In addition, an earlier study, in Egypt (Tiller et al., 2007) applied the MLVA-15 method for genotyping analysis for B. melitensis strains of human origin. While, in a recent study, (Abdel-Hamid et al., 2020) showed that B. abortus strains from Egypt are closely linked to clonal lineages of the Western, Mediterranean, and East Asian and Americas strains as well as B. melitensis strains were similar in their familiarity. On the other hand, these findings were not in touch with (Sayour et al., 2020) who examined 118 Egyptian B. melitensis bv3 strains by MLVA-16 that typing these strains onto 70 genotypes, 51 of them produce high genetic diversity with the West Mediterranean clones. The high homology and similarities of M. bovis strains in our study with various regional strains from multiple foreign strains. While this not agree with (Register et al., 2015) who reported high distinct diversity among M. bovis strains from cattle and bison indicated the specific sets of sequence types by MLST method. Furthermore, sequences of uvrC, gapA and p40 pseudo gene of bovine M. bovis strains from pneumonia in Egypt was demonstrated by (Abdeen et al., 2017) and displayed strong humongous and similarity with other local and global strains. Furthermore (Amram et al., 2013) used MLVA method to distinguish eleven M. bovis genotypes strains from other strains. Interestingly, several recent studies have demonstrated the great variety of M. bovis strains among different animal species; for example, (Marianelli et al., 2019) recognized 73 distinct genetic genotypes among 14 small-scale herds and 21 diverse regions of Messina province in Italy. Next, we analyzed a single strain of L. monocytogens from aborted cow using the 16SrRNA gene that demonstrated high identity and homology (99.78-100%) on the gene bank with local and multiple nation strains. This exploited the 16S rRNA gene for phylogenetic analysis was in connection with (Soni and Dubey, 2014) as well as (Soni et al., 2014) reported the close association more than 99% between the members of Listeria species. Also, (Konstantinidis et al., 2007) indicated that a 16S rRNA diversity greater than 5 % or identity lower than 70 % in amino acid sequence could be used as a tool to distinguish between the genus listeria. Moreover, (Qin et al., 2014) reported that the well-stable proteins more than 50 % should be used as a threshold level to describe the species in the same community. According to (CFSPH, 2019) the nucleotide sequencing and MLST technique are specific alternative methods for distinguishing the clonal complex of L. monocytogenes strains as well as the important for the general and phylogeny characters among various listeria strains in population.
In conclusion, Abortion is still a major devastating problem in all dairy herds caused by various bacterial causes as Brucella, Listeria, and Mycoplasma species. This study focused supremacy of B. abortus, B. melitensis, L. moncytogenes and M. bovis as prime causes of cattle abortion in Egypt. In addition to the great specificity and sensitivity of PCR method for efficient identification of theses bacteria from aborted cases. Moreover, the sequencing analysis revealed the identical and high genetic similarities between our strains and other forging strains of different sources indicating the distribution of these strains between different countries and localities during the animal movement and importation. Also, this study emphasized the role application of hygienic measures and vaccination programs for control of infectious diseases to avoid the dissemination of infectious agents among cattle.
Acknowledgements
The authors would like to thank the staff members of Animal Medicine and Infectious Diseases, Faculty of Veterinary Medicine, University of Sadat City as well as Animal Health Research Institute, Agricultural Research Center for the help for finishing the manuscript.
Conflict of interest
The authors have no conflict of interest.
Authors contributions
W.S.M., E.E. A., M.A.N. and Y.H. involved in the conception of the research idea and methodology design, performed data analysis and interpretation, and prepared the manuscript for publication, H.T. E participated in the design of the methodology, involved in laboratory work and H.T. E, E.I.B, S. M.S. involved in laboratory work and data analysis and contributed their scientific advice during the work and revision. All authors read and approved the final manuscript.
References






