Advances in Animal and Veterinary Sciences
Research Article
Antibiotic Resistance of Escherichia Coli in Pork Sold at Tamiang Layang Market, East Barito District
Akhmad Rizaldi1, Denny Widaya Lukman2*, Herwin Pisestyani2
¹Graduate School, Study Programs of Veterinary Public Health, IPB University; ²Department of Animal Infectious Diseases and Veterinary Public Health, Faculty of Veterinary Medicine, IPB University, West Java, Indonesia 16680.
Abstract | This study aimed to determine the occurrence of antibiotic resistance of Escherichia coli (E. coli) isolated from pork sold inTamiang Layang Market, East Barito District. We took a total of 41 upper thigh pork samples were taken from all of the Tamiang Layang Market pork traders. Isolation and identification of E. coli were carried out using Brilliance E. coli/Coliform selective Agar, Eosin Methylene Blue Agar (EMBA), and confirmed with Analytical Profile Index (API) 20E. The resistance against ten antibiotics (nalidixic acid, erythromycin, ampicillin, streptomycin, penicillin G, sulfamethoxazole, ciprofloxacin, tetracycline, amoxicillin, and chloramphenicol) were tested using Kirby Bauer method, based on the standard of Clinical Laboratory Standards Institute (CLSI) in 2012. The result showed that 17 samples were positive containing E. coli (41.5%) and they all resistant to erythromycin, streptomycin, penicillin G, and chloramphenicol (100%), tetracycline (94.1%), ampicillin (76.5%), and nalidixic acid (23.5%). All isolates were also resistant to more three classes of antibiotics which were known as Multi-Drug Resistant (MDR) with mostly pattern of E-AMP-S-P-TE-C (erythromycin, ampicillin, streptomycin, penicillin G, tetracycline, and chloramphenicol).
Keywords | Antibiotic resistant, E. coli, Multi drug resistant, Pork
Received | March 31, 2018; Accepted | May 01, 2019; Published | August 26, 2019
*Correspondence | Denny Widaya Lukman, Department of Animal Infectious Diseases and Veterinary Public Health, Faculty of Veterinary Medicine, IPB University, West Java, Indonesia 16680; Email: dennylukman@hotmail.com
Citation | Rizaldi A, Lukman DW, Pisestyani H (2019). Antibiotic resistance of escherichia coli in pork sold at tamiang layang market, east barito district. Adv. Anim. Vet. Sci. 7(9): 791-797.
DOI | http://dx.doi.org/10.17582/journal.aavs/2019/7.9.791.797
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2019 Rizaldi et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
Pig is well known as the primary livestock raised by the people in the East Barito District. The majority of East Barito communities are from Dayak tribes who use pork in traditional ceremonies. They also like to raise pigs because the maintenance costs are relatively cheap; the growth of the body is relatively fast and does not require extensive maintenance cages. Pigs, well known as omnivores, can consume agricultural wastes as sources of their feed. Also, pigs have a high litter size, and people often use their manure to increase soil fertility. Pork is one of the essential commodities regarding the aspect of its nutrient, socio-culture, and economy (Priadi et al., 2016).
In 2016, the population of pig farms in the East Barito District reached 36 667 pigs, and about 7 514 pigs were slaughtered (BPS Barito Timur, 2017). Hence, pig farms provide a promising prospect for the people of East Barito. Unfortunately, many traditional markets in East Barito do not yet have special kiosks for the sale of pork. Currently, pork in East Barito sold on the roadside of the Tamiang Layang Market. The condition can cause the pork directly exposed to the sun, rain, dust, and other contaminants from the highway, which further may affect the pork quality.
Food that sold openly on the roadside has a higher chance of being contaminated by coliform bacteria and pathogenic bacteria such as Escherichia coli, Salmonella sp., Staphylococcus aureus, Bacillus cereus, Clostridium perfringens, and Vibrio cholerae (Cho et al., 2011; Hanashiro et al., 2005; Mankee et al., 2005). Contamination of pathogenic bacteria in meat may result in foodborne disease (CDC 2018).
Escherichia coli (E. coli) is a pathogenic bacteria causing foodborne disease that is harmful to human health. E. coli is also known as commensal bacteria which commonly used as an indicator of the sanitation level and microbial resistance on livestock and its products. Commensal bacteria that is resistant against antibiotics will serve as a reservoir in spreading resilient characteristic to other bacteria, both in animal and human, through the food chain or direct contact (Sari, 2018). Escherichia coli can produce Shiga toxin, known as Shiga toxin-producing Escherichia coli (STEC) which causes foodborne disease. The primary source of STEC disease includes raw meat, raw milk, and fecal contamination of vegetables (WHO, 2018).
Escherichia coli (E. coli) is a pathogenic bacteria causing foodborne disease that is harmful to human health. E. coli is also known as commensal bacteria which commonly used as an indicator of the sanitation level and microbial resistance on livestock and its products. Commensal bacteria that is resistant against antibiotics will serve as a reservoir in spreading resilient characteristic to other bacteria, both in animal and human, through the food chain or direct contact (Sari, 2018). Escherichia coli can produce Shiga toxin, known as Shiga toxin-producing Escherichia coli (STEC) which causes foodborne disease. The primary source of STEC disease includes raw meat, raw milk, and fecal contamination of vegetables (WHO, 2018).
Bacteria that are resistant to antibiotic become a new problem nowadays as they may cause infectious disease that is difficult to treat and requires higher medical expenses. Antibiotic-resistant pathogenic bacteria have been growing significantly and infecting human and animal (Kallau et al., 2018). This resistance also occurs in bacteria found in pork meat.
Hammerum and Heuer (2009) mentioned that E. coli from animals could perform as a donor of antibiotic resistance genes to other pathogenic E. coli. According to Liu et al. (2016), food contaminated with antibiotic-resistant bacteria consumed by humans and animals may lead to the growth of resistant bacteria both in humans and animals. Study on sanitation related to contamination of antibiotic resistant E. coli in pork is still limited especially in Indonesia. Therefore this research was conducted to observe the occurrence of antibiotic-resistance E. coli in pork sold in Tamiang Layang Market, East Barito District.
METHODS
Sampling Method
About 250 grams of pork meat samples were collected aseptically from the upper part of the thigh from all pork stalls (41 samples) in Tamiang Layang Market. We took three times sampling at different days. A small number of samples used in this study because there are only 8 pork traders in the Tamiang Layang Market. The samples collected at 07.00-09.00 a.m. Samples were put into sterile plastic bags and labeled according to sampling location, stored in a cool box with a temperature of 4-10 ºC and immediately taken to the Laboratory of Veterinary Public Health of Regional Disease Investigation Center (BVet) of Banjarbaru for isolation test and identification of E. coli. We also examined the resistance test against ten selected antibiotics (nalidixic acid, erythromycin, ampicillin, streptomycin, penicillin G, sulfamethoxazole, ciprofloxacin, tetracycline, amoxicillin, and chloramphenicol).
Isolation and Identification of Escherichia coli (E. coli)
Escherichia coli isolation and identification test were referred to the guideline for laboratory analysis on an examination of microbial contamination in meat, egg, and milk according to SNI 2897:2008 and working instruction of SNI ISO/IEC 17025:2008 from the National Standardization Agency of Indonesia (BSN). American Type Culture Collection (ATCC) 25922 E. coli isolates were used as positive controls in each test conducted. We weighted about ten grams of pork sample and added with 90 ml 0.1% Buffered Peptone Water (BPW). After that, the mixture was homogenized using stomacher for 1 minute, put into Erlenmeyer flask and then incubated at a temperature of 41.5 ºC for 6 hours. Thus, the inoculation was conducted by transferring one inoculating loop of the sample to selective medium brilliance E. coli/Coliform agar. The agar incubated at a temperature of 35 ºC for 18-24 hours. On this brilliance
E. coli/Coliform selective agar, E. coli colony showed purplish-blue/dark blue color. The expected E. coli colony was then inoculated to Eosine Methylene Blue Agar (EMBA) medium that is a selective stain for gram negative bacteria. The colony was incubated at a temperature of 35 ºC for 18-24 hours. On EMBA medium, the colony expected to be E. coli showed black/dark color, while the center of the colony was metallic green. The colony expected to be E. coli on EMBA medium was then inoculated to blood agar media to observe hemolytic characteristic. On this blood agar medium, the expected colony of E. coli was inoculated twice to obtain a pure culture. Gram staining was performed on the expected E. coli colony from blood agar followed by inoculation on MacConkey agar, oxidase test, Methyl Red (MR) test, and motility test. The colony with a positive result was cultured on Nutrition Agar (NA) slant. After that, we incubated the colony at a temperature of 35 ºC for 18-24 hours. Isolates from NA slant then confirmed using kit API 20E (Biomereaux). Lastly, Apiweb TM application was used to read the result.
Antibiotic Resistance Test
Resistance test conducted was based on the standard of Clinical and Laboratory Standards Institute (CLSI) in 2012. We used CLSI 2012 because this study was conducted in the Banjarbaru Veterinary Center laboratory. In Banjarbaru Veterinary Center laboratory, the method used according to the CLSI 2012. We perform the antibiotic resistance test to all E. coli colonies obtained from the isolation of pork samples. Bacteria prepared in the form of suspension equivalent to 0.5 McFarland turbidity standard (1-2x108 CFU/mL). The culture was taken using a sterile cotton swab, spread on Mueller Hinton Agar (MHA) surface, and left for ± 5 minutes. Later, with applied Kirby Bauer method, the commercial paper disk contained antibiotic was put on MHA, which was previously spread by the pure culture at distance of 25-30 mm. The culture was incubated at a temperature of 35 ºC for 18-24 hours. Determination of susceptible, intermediate, and resistant categories was done based on the size of the zone of inhibition formed according to the standard of CLSI 2012. A blank disk without antibiotic was used as negative control for each test.
Data Analysis
Data were analyzed descriptively in the form of figures.
RESULTS
The result showed that 17 samples were positive containing
E. coli (41.5%) (Figure 1) and they all resistant to erythromycin, streptomycin, penicillin G, and chloramphenicol (100%), tetracycline (94.1%), ampicillin (76.5%), and nalidixic acid (23.5%). However, the isolates were still susceptible to sulfamethoxazole, ciprofloxacin, and amoxicillin (Figure 2). All isolates were also resistant to more three classes of antibiotics, known as Multi-Drug Resistant (MDR), with mostly pattern of E-AMP-S-P-TE-C (erythromycin, ampicillin, streptomycin, penicillin G, tetracycline, and chloramphenicol).
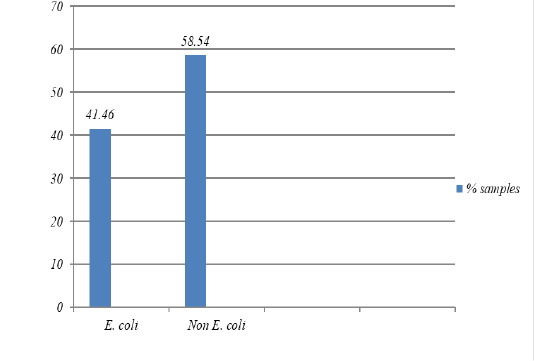
Figure 1: The percentage of isolates of bacteria E. coli was isolated from pork Market in the Tamiang Layang, East Barito Regency
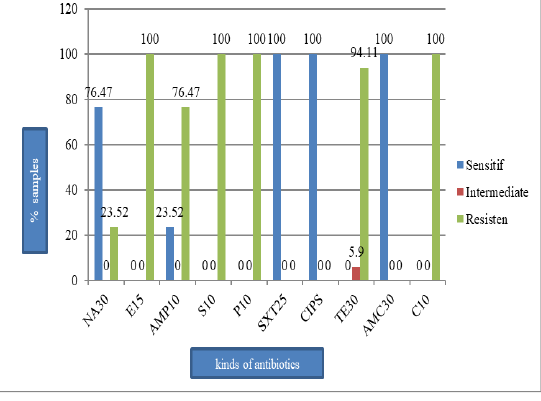
Figure 2: The percentage of E. coli sensitive, intermediate, and resistant on the pork sold in Tamiang Layang Market; nalidiksat acid (NA30), erythromycin (E15), ampisilin (AMP10), streptomycin (S10), penicillin G (P10), sulfametasole (SXT250), ciprofloxacin (CIPS), tetracycline (TET30), amoksilin (AMC30), and chloramphenicol (C10).
DISCUSSION
The Result of the Isolation Test and Identification of E. coli
Out of the total 41 pork samples collected from the upper part of the ham and tested in this study, 17 samples (41.5%) were contaminated with E. coli. The result is in agreement with the study conducted by Adesiji et al. (2011), showed that there was 40% of E. coli contamination in pork meat from Taiwan pork samples.
Escherichia coli is a cosmopolite bacteria mainly found in the environment. The species can live in an aerobic or anaerobic condition and survive in nutrient-poor media such as water, floor, and inorganic surface (Bell and Kyriakides 2002). In a slaughterhouse, E. coli contamination may result from animal feces or offal disposal process. Feces containing E. coli will contaminate equipment used for slaughtering, floors, and environment around the slaughterhouse. In this study, the sellers in Tamiang Layang Market obtained pork that was not slaughtered in a particular pig slaughterhouse. They traditionally slaughter animals in their house or other places around the market. So the possibility of cross-contamination by feces or other sources.
The pork kiosks at Tamiang Layang Market are located precisely on the edge of the road. Other than that, the sellers sold pork in the kiosks openly without any cover. Food sold openly on the roadside has a high chance of contamination of coliform bacteria and pathogenic bacteria such as Escherichia coli, Salmonella sp., Staphylococcus aureus, Bacillus cereus, Clostridium perfringens, and Vibrio cholerae (Cho et al., 2011; Hanashiro et al., 2005; Mankee et al., 2005). Contamination can occur through the air, flies, the hands of the buyer, the meat mat, and from the equipment used when selling. The air is primarily a medium for the spread of microorganisms. The group of organisms spread in free air is bacteria and fungi (Waluyo, 2005). Susanna et al. (2010) stated that flies that perch on food products would contaminate these food products.
Bacteria of E. coli can be used either as an indicator of sanitation for food product quality or to observe the fecal contamination during its production stage (Susanto, 2014). The presence of E. coli in food products is related to the existence of other pathogenic bacteria. OIE (2013) explained that monitoring of antibiotic resistance could be done with the help of indicator bacteria such as E. coli from the animals, animal food products, and humans.
The Result of Antibiotic Resistance Test
This study showed that 17 E. coli isolates tested resistant to antibiotics as follows: 100% were resistant to erythromycin, penicillin G, and chloramphenicol; 94.1% were resistant to tetracycline; 76.5% were resistant to ampicillin, and 23.5% were resistant to nalidixic acid. However, the isolates were still susceptible to sulfamethoxazole, ciprofloxacin, and amoxicillin. The result of this study is in agreement with the result found by Azizah et al. (2012) that 100% of E. coli isolates from pig were resistant to chloramphenicol.
In Taiwan, Hsu et al. (2006) found that E. coli isolates from pork were 100% resistant to tetracycline and streptomycin, 93.4% resistant to chloramphenicol and ampicillin, and 95.1% resistant to nalidixic acid. In Korea, Lim et al. (2007) found that E. coli isolated from the pig was resistant to tetracycline (96.3%), streptomycin (66.8%), ampicillin (66.1%), chloramphenicol (47.6%), sulfamethoxazole (38.8%), and ciprofloxacin (7.8%).
The incidence of antibiotic resistance that frequently occurred over the last few years was caused by imprudent use of antibiotics for treatment in human and animal (Barton 2000). Antibiotics are delivered to animals for a variety of reasons, including disease treatment, prevention, control, and growth promoters to increase livestock productivity. Antibiotic from the group of macrolide, polypeptide, phenicol, and aminoglycoside was used in the USA to boost livestock growth during the mid-1990s (Marshall and Levy 2011).
According to Harada and Asai (2010), many livestock farmers in Japan used 870 tons of antibiotic from the group of tetracycline, chloramphenicol, aminoglycoside, fluoroquinolone, and sulfonamide for poultry, pig, and cattle. In pig farms located in Solo, about 14 types of antibiotics were used for pig disease treatment or prevention. Type of antibiotic mainly used by a pig farmer in Solo consisted of penicillin, sodium sulfadimethylpyrimidine, oxytetracycline, and amoxicillin (Arief et al., 2016).
According to the result of interview and observation with animal health officers and pig farmers in East Barito District, it was found that animal health officer usually used oxytetracycline, tetracycline, and amoxicillin (100%) for treatment of pigs. Around 44.4% of pig farmers were found to apply antibiotics used in human medicine in the treatment of pig, such as ampicillin, amoxicillin, and kanamycin. One pig farmer (3.7%) who mixed antibiotic in drinking water or animal feed.
Some isolates were found to be resistant toward the antibiotic group of beta-lactam (penicillin and ampicillin) due to the ability of E. coli bacteria to produce beta-lactamase enzyme that hydrolyzes the beta-lactam ring of the antibiotic compound. In the antibiotic group of chloramphenicol, resistance may exist due to the ability of bacteria to produce chloramphenicol acetyltransferase (CAT) enzyme that encodes the resistant gene of chloramphenicol (Boerlin and White 2006). In a group of aminoglycoside (streptomycin), resistance occurs because bacterial cells were able to inactivate antibiotic and modify antibiotic target (Guifole, 2007).
Kallau et al. (2018) mentioned that erythromycin ribosome methylation (ERM) is the gene responsible for erythromycin resistance, which usually occurs in pig farms. In the tetracycline group, resistance exists due to changes in the ribosome and multidrug efflux pump process (Sen and Sarkar, 2018). Frequent and long-term use of tetracycline in the field will lead to tetracycline resistance (Dai et al., 2008).
Animal health officers and pigs’ farmers in East Barito did not use chloramphenicol for the treatment of pig diseases. Azizah et al. (2002) found a rare use of chloramphenicol treatment because it may lead to hypersensitive reaction, anorexia, and stimulate the development of teratogenic characteristics. Hamscher et al. (2003) found that out of 90% dust samples from bedding, feed, and feces in pig farms in Germany contained 12.5 mg/kg of the antibiotic residue of tylosin, tetracycline, sulfamethoxazole, and chloramphenicol.
Animal health officers and pig farms did not use erythromycin and streptomycin in Tamiang Layang Market. The two types of antibiotic used frequently in commercial poultry because of their broad spectra. Location of commercial poultry that is close to the river in East Barito District may trigger the spread of resistant E. coli bacteria to the residential area and the environment around the poultry farms. This antibiotics resistance spread to humans and animals directly by contact and indirectly via the food chain, water, air, and manures (Marshall and Levy, 2011).
Manures has become a reservoir of resistant bacteria and antibiotics compounds, and its applications to agricultural soils are assumed to increase antibiotics resistance genes in soil significantly (Heuer et al., 2011). A study conducted by Jiang et al. (2011) indicated that antibiotic-resistant E. coli bacteria from poultry, pig, and dairy cattle farms can easily be found in the sample of water and soil around the farm area. An aquatic environment such as water and sediment plays an essential role in the transfer and evolution of antibiotic resistant gene (Marti et al., 2014).
Multi-Drug Resistance (MDR) Escherichia coli
Multi-drug resistance (MDR) is the condition where bacteria are resistant to three or more classes/groups of antibiotic (Magiorakos et al., 2012). In this study, seventeen isolates of E. coli classified as MDR with the most pattern of
E-AMP-S-P-TE-C (erythromycin, ampicillin, streptomycin, penicillin G, tetracycline, and chloramphenicol). The finding reported from this result in an agreement to the result of a study performed by Hsu et al. (2016) in Taiwan that found that all E. coli isolates obtained from pork meat has been identified as MDR. The MDR pattern found in Taiwan was AMP-CB-TET-STR-GEN-KAN-SPT-CHL-ERY-SU-NAL (ampicillin, carboxypenicillin, tetracycline, streptomycin, gentamycin, kanamycin, spectinomycin, chloramphenicol, erythromycin, sulfonamide, and nalidixic acid).
Lim et al. (2007) found that all E. coli isolates originated from the pig in Korea has become MDR with the most frequent pattern of AM-S-C-TE (amoxicillin, streptomycin, chloramphenicol, and tetracycline). In Indonesia, Kallau et al. (2018) found that 47 of 82 E. coli isolates from pig farms in Kupang City were MDR with the most frequent pattern of KF-CT-E (cephalothin, colistin, and erythromycin). Friendship (2006) also mentioned that E. coli and Salmonella found in pork were resistant to several antibiotics. Krisnaningsih et al. (2005) reported that in every bacterial resistance case, multidrug resistance such as ampicillin (the derivative of penicillin), streptomycin, and tetracycline was often found, especially concerning E. coli and Salmonella.
High incidence of MDR E. coli found in pork sold in Tamianglayang market requires special attention, particularly from related institution since it may lead to public health problem in the future. High incidence of MDR in bacteria is closely related to the ability of bacteria to transfer resistant gene through spontaneous DNA mutation, transformation, and pass through by plasmid as an intermediary. Jakobsen et al. (2010) found that all E. coli isolates obtained from pork meat in Denmark were MDR. The incidence of MDR such as ampicillin, streptomycin, sulfamethoxazole, tetracycline, and chloramphenicol was related to transfer horizontally of chloramphenicol resistance genes. A similar finding was also confirmed by Bischoff et al. (2005) who found that cmlA gene against chloramphenicol was able to share the resistant characteristic through plasmid transfer, causing the existence of MDR E. coli in pig farms.
CONCLUSION
This study showed that 17 samples (41.5%) were positive to contain E. coli of 41 pork samples collected in Tamiang Layang Market, East Barito District. Seventeen isolates of E. coli obtained were 100% resistant, against erythromycin, streptomycin, penicillin G, and chloramphenicol. Furthermore, those seventeen isolates of E. coli identified as multidrug-resistant bacteria with mostly pattern of E-AMP-S-P-TE-C (erythromycin-ampicillin-streptomycin-penicillin-tetracycline-chloramphenicol).
ACKNOWLEDGMENT
This research was support by the scholarship from East Barito District, Central Kalimantan Province, Indonesia.
CONFLICT OF INTEREST
There is no conflict of interest in this review to declare.
AUTHORS CONTRIBUTION
The research was designed jointly by Akhmad Rizaldi, Denny Widaya Lukman, and Herwin Pisestyani. Akhmad Rizaldi conducted the research. The authors have read and approved the final manuscript.
REFERENCES
Nutr. Res. Rev. 13(2):1-19.
Env. Mic.18:557-564.






