Advances in Animal and Veterinary Sciences
Research Article
Molecular detection of Salmonella typhimurium isolated from canine feces by PCR
Worod Salih, Afaf Abdulrahman Yousif*
University of Baghdad, College of Veterinary Medicine, Al Gaderia, Iraq.
Abstract | This study was conducted to detect five virulence factors include invA, spvC, fimA, stn and Typh genes on 4 isolates of S. typhimurium serotype which isolated previously from 3 puppies and 1 adult dog, this isolates were confirmed by culture, biochemical, Gram stain and serotyped at the National Center of salmonella in the Baghdad / Ministry of Health. The 4 isolates [3 isolated from puppies and 1 from adult] were confirmed by conventional PCR, A five genes were designed for molecular study, these include invA, spvC, fimA, stn and Typh genes. The results of molecular study showed that all isolates were positive to Typh gene, while invA gene was detected in two isolates only. One isolate only was having SpvC gene. The results of fimA gene showed presence of this gene in two isolates, while the Stn gene was not detected in in any isolates of S. typhimurium. In conclusion, the Salmonella typhimurium had been detected in dogs and puppies which can cause an infection in human and PCR system is useful in the detection of virulence of this bacteria.
Keywords | Salmonella, Molecular, Canine, S. typhimurium, Gene
Editor | Kuldeep Dhama, Indian Veterinary Research Institute, Uttar Pradesh, India.
Received | August 08, 2018; Accepted | October 04, 2018; Published | October 18, 2018
*Correspondence | Afaf Abdulrahman Yousif, University of Baghdad, College of Veterinary Medicine, Al Gaderia, Iraq; Email: afaf_a.rahman@yahoo.com
Citation | Salih W, Yousif AA (2018). Molecular detection of Salmonella typhimurium isolated from canine feces by pcr. Adv. Anim. Vet. Sci. 6(12): 542-547.
DOI | http://dx.doi.org/10.17582/journal.aavs/2018/6.12.542.547
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2018 Salih and Yousif. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Salmonella spp. remains a significant food borne zoonotic pathogen in all regions of the world with S. typhimurium as one of the most common serovars causing disease in animals and human (Herrero-Fresno and Olsen, 2018).
Salmonella is gram-negative, flagellated, non-encapsulated, non– sporulated and aerobic (facultative aerobic) bacilli, they are usually seen as short rods and occasionally develop into a longer pleomorphic form or short coccobacilli when incubated for a long time in the laboratory (Markey et al., 2013).
Salmonella can cause a variety of pathologies, including gastroenteritis, abortions, pneumonia and lethal septicemia in human and animals (Ohl and Miller, 2001). Dogs generally seem to be resistant to Salmonella infection and most cases are latent and non-clinical, infected dogs may remain carriers and faecal shedders and thus serve as sources of Salmonella for man and other animals (Kozak et al., 2003).
A study of Verma et al. (2011) on clinical cases of canine salmonellosis was designed to compare the sensitivity, specificity and accuracy of PCR to the isolation and characterization methods commonly used in diagnostic laboratories, they indicate that the PCR assay as a simple, rapid, sensitive, reliable and reproducible method for the identification of Salmonella that will aid in surveillance, prevention and control of this pathogen.
Makino et al. (1999) established the specific detection to Salmonella species by PCR system using Salmonella enterotoxin gene (stn),This detection was limited to one bacterial cell/ one gram of fecal samples and a minced-meat using enrichment by Salmonella enrichment broth or Tripticase soy broth respectively, They concluded that PCR system is useful in the field of the public hygiene.
This study aimed to detect the virulence factors in Salmonella typhimurium isolated from canine feces.
Materials and Methods
Bacterial Isolates
Four S. typhimurium were isolated in pervious study (Ibrahim and Yousif, 2018), 3 isolates from puppies with different clinical signs and one isolate from adult dog, these isolates were conducted to cultural, biochemical in the dept. of Internal and Preventive Vet. Medicine and confirmed serologically as S. typhimurium in the national center of Salmonella in Baghdad.
Table 1: Primers types with their sequences with product size
| Primers | Primer sequence (5- to 3- ) | Product size (bp) | References | |
| invA | F | GTG AAA TTA TCG CCA CGT TCG GGC AA | 284 |
Kumar et al.(2008) |
| R | TCA TCG CAC CGT CAA AGG AAC C | |||
| spvC | F | ACT CCT TGC ACA ACC AAA TGC GGA | 571 |
Oliveira et al.(2003) |
| R | TGT CTT CTG CAT TTC GCC ACC ATC A | |||
| fimA |
F
|
CCT TTC TCC ATC GTC CTG AA | 85 |
Naravaneniet al.(2005) |
| R | TGG TGT TAT CTG CCT GAC CA | |||
|
F
|
CTT TGG TCG TAA AAT AAG GCG | 260 |
Makino et al.(1999) |
|
| R | TGC CCA AAG CAG AGA GAT TC | |||
| Typh |
F
|
TTG TTC ACT TTT TAC CCC TGA A | 401 |
Olsen et al., 1995 |
| R |
CCC TGA CAG CCG TTA GAT ATT |
|||
Table 2: Number of isolates that carried virulence genes in affected animals
| Animals (4) | No. of S. typhimurium isolates(4) |
|
Gene | Total of genes in each isolates | |||
| Typh | fimA | SpvC | InvA | Stn | |||
|
1- Puppy with bloody diarrhea
|
1 | | |
|
|
- | 4 |
| 2-Puppy with diarrhea | 2 | | | - | 2 | ||
| 3-Non diarrheic Puppy | 3 | | - | 1 | |||
| 4-Adult dog with bloody diarrhea | 4 | | | - | 2 | ||
Molecular Study in Isolated Bacteria by Conventional Pcr
Four isolates of Salmonella typhimurium were used in the molecular study, these isolates were subjected:
DNA extraction: According to manufacturing procedure. The DNA of Salmonella isolates was extracted by using by using (Geneaid DNA Bacterial Kit, USA).
Primers: Five primers in this study were purchased from Bioneer, Koreato detect salmonella virulence gene. These primers were prepared according to the information of the company. invA, spvC, fimA, stn and Typh genes (Table 1).
The concentration and purity of extracting DNA were evaluatedby using a Nanodrop (ND-2000,Thermo Fisher scientific, USA). Then gel electrophoresis was used to check the extracted DNA by loading the DNA in 1.5% agarose gel.
PCR mixture component of invA, spvC, fimA, stn and Typh genes: For detecting invA, spvC, fimA, stn and Typh genes of Salmonella typhimurium by PCR, the PCR amplification mixture which used for detection these genes includes master mix (12.5μl), 2 μl of template DNA, 2μl of each forwarded and reversed primers and 6.5μl of nuclease free water to complete the amplification mixture to 25μl.
Thermo- cycler program: The program of thermo-cycle for detection invA, Typh and SpvC genes as follows:
One cycle for 3 minutes at 94°C to denature template. It was continued by 35 cycles, each cycle including denaturation for 30 seconds at 94°C, annealing 30 seconds at 63°C, and extension 30 seconds at 72°C. Final extension was done 5 minutes at 72°C.
The program of thermo-cycle for detection of fimA gene was done as follows: Initial denaturation ongoing by 1 cycle at 94°C for 3 min. Followed by 35 cycles, each cycle including denaturation, annealing and extension at 94°C for 30 seconds, 56°C for 30 sec., 72°C for 30 sec. The 35 cycles Followed by a final extension for 5 min. at 72°C.
The amplification of stn gene was carried out employing same conditions as invA, Typh and spvC genes except annealing at 55°C.
PCR Product Analysis
This step was used for analyzing the PCR product by using 2% agarose, stained with 0.5 μg/mL ethedium bromide, The PCR products (bands) were visualized by using a UV transilluminator and photographed by using digital camera.
Results
DNA genomic extraction of salmonella
The four isolates of Salmonella were showed compact bands as an indication of DNA extraction successfully in gel electrophoresis (Figure 1).
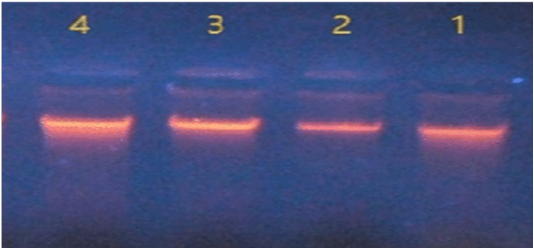
Figure 1: DNA extraction of Salmonella isolates.
In Nano drop spectrophotometer the concentration of DNA ranged between 47 – 190 ng/μl and the purity were between 1.6 – 2.1 at absorbance 260/280 nm.
Confirmatory Detection of Virulence Genes by Using Conventional Pcr
The results of PCR amplification of the extracted DNA from isolates of S. typhimurium on different types of genes used in comparison with DNA marker (100bp ladder) as follows.
Typh gene was amplified DNA at 401bp fragments , All isolates were positive for this gene (Figure 2). While invA gene was detected in two isolates only which revealed 284bp (Figure 3).
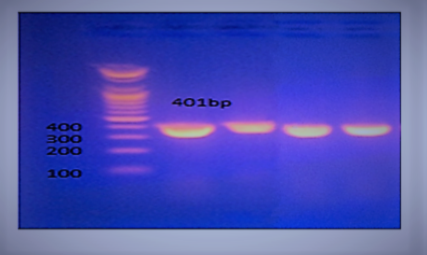
Figure 2: Agarose gel electrophoresis showing amplification of 401 bp fragments of Typh gene. Lanes 1,2,3,4 showing positive amplification of Salmonella typhimurium
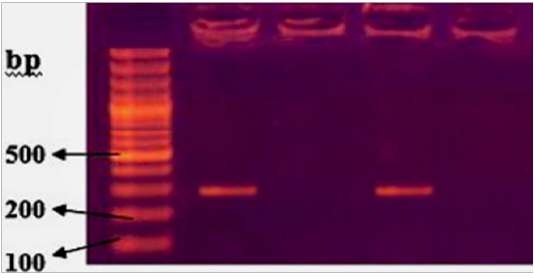
Figure 3: Agarose gel electrophoresis showing amplification of 284bp fragments of invA gene. Lanes 1,3 showing positive amplification of Salmonella typhimurium

Figure 4: Agarose gel electrophoresis showing amplification of 571bp fragments of SpvC gene. Lane 1 showing positive amplification of Salmonella typhimurium.
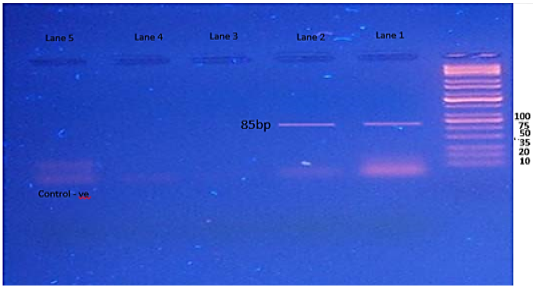
Figure 5: Agarose gel electrophoresis showing amplification of 85bp fragments of fimA gene. Lanes 1,2 showing positive amplification of Salmonella typhimurium.

Figure 6: Agarose gel electrophoresis showing amplification of 280bp fragments of stn gene. Lanes 1,2,3,4 showing negative amplification of Salmonella typhimurium.
The SpvC gene showed amplification of 571bp fragments, one isolate only was have this gene (Figure 4). The results of fimA gene showed amplification of 85bp fragments in two isolates of S. typhimurium (Figure 5), while the Stn gene was not detected in in any isolates of S. typhimurium. (Figure 6).
Distribution Of Virulence Genes Among Infected Animals
The presented results in a Table 2 revealed the distribution of different genes in each isolate of Salmonella typhimurium. The first isolate from Puppy with bloody diarrhea carried 4 genes (Typh, fimA, SpvC & InvA), second isolate from -Puppy with diarrhea carried 2 genes (Typh & fimA), the third isolate from-Non diarrheic Puppy was carried Typh gene only. While the Salmonella isolate from adult dog was possessed 2 genes (Typh, & InvA).
Discussion
All the results of culturing, gram stain and biochemical tests are similar to the morphological characters were compatible with the information mentioned by (Markey et al., 2013; Mahon et al., 2015).
In-vitro amplification of DNA by the PCR method is a powerful tool in microbiological diagnostics (Malorny et al., 2003). Several genes have been used to detect Salmonella in natural environmental samples as well as food and faecal samples.
The current result showed that the PCR was a sensitive and specific method as compared with bacterial culture and this result was in agreement with a result of Kurowski et al. (2002) who used PCR technology to develop a highly sensitive and specific diagnostic assay for the detection of Salmonella spp in fecal dogs specimens, they found that Use of the spaQ primer-probe set resulted in a relative sensitivity of 100% and a specificity of 98.2%, compared with bacterial culture results when tested on 299 clinical fecal specimens.
Also, the results were incompatible with result of Parungao et al. (2010) who used conventional isolation procedures and PCR for the presence of Salmonella spp. in feces of dog, they detected salmonella in both conventional microbiological procedures (1 out of 62 or 1.6%) and PCR (26 out of 62 or 42%) of the samples tested, they concluded that apparently healthy dogs can be carriers and shedders of potentially zoonotic salmonellae, and that PCR can be an effective means of detecting the said pathogen in canine fecal samples.
Virulence chromosomal genes including, invA, invE, himA, phoP are target genes for PCR amplification of Salmonella species (Jamshidi et al., 2009).
The invA gene of Salmonella contains sequences unique to this genus and has been proven as a suitable PCR target, with potential diagnostic applications (Rahn et al., 1992). Amplification of this gene now has been recognized as an international standard for detection of Salmonella genus (Malorny et al., 2003).
The presented results showed a high prevalence of invA gene at percent 50%, this result agree with result of Hashemi et al. (2014) who aimed to find out the prevalence rate of Salmonella infection in apparently healthy cats and dogs in Iran by using PCR by using invA gene, they found that prevalence rates of Salmonella were 18 and 22% in cats and dogs respectively, also incompatible with Srisanga et al. (2017) who showed that most canine Salmonella isolates harbored invA (100%), and agree with Torkan et al. (2015) who reported a high prevalence of invA gene in Salmonella isolates, this result indicates that pet dogs and cats serve as reservoirs of invasive Salmonella.
Also, Chaudhary et al. (2015) isolate Salmonella Enteritidis and Typhimurium from pork and slaughter house environment in Ahmedabad, Gujarat out of a total of 270 samples, 37 (13.70%), All Salmonella serovars produced invA gene, fimA and amplicon for enterotoxin (stn) gene whereas 30 isolates possessed 310 bpspvR gene, but no isolate possessed spvC gene.
Kshirsagar et al. (2014) found a high percent invA, stn and fimA genes associated with Salmonella isolated from the retail meat market of raw buffalo meat and offals viz. liver, lung, muscle, intestine and ground beef in Gujarat India. In Iraq, Al-Zubaidy et al. (2015) detected the invA gene in the Salmonella spp. isolated from the cows
A total of 37 Salmonella isolates of 11 different serotypes and rough type of human and animal clinical cases and meat samples were studied for the presence of 8 virulence determinants including 4 virulence genes and 4 toxic factors, All Salmonella isolates harbored invA and stn genes (Singh et al., 2013), these results were didn’t agree with our result about the stn gene which not presented in the current study. Ulaya (2013) showed that the invF gene was (16.7%) as well as the sipC gene (8%) the hilD (5.6%) gene belonging to Salmonella in Zambia from dogs.
Conclusion
Salmonella isolated from companion animals (dogs and puppies) and these have a many virulence factors which can transmit to human and causing infection.
ACKNOWLEDGMENTS
This work was supported by College of Veterinary Medicine, Department of Internal and Preventive Veterinary Medicine, University of Baghdad and Ministry of agriculture / Iraq.
Conflict of interest
The author declares that they have no competing interests.
Authors Contribution
All the authors contributed equally.
References






