Advances in Animal and Veterinary Sciences
Research Article
Predominance of G10 Genotype of Rotavirus in Diarrheic Buffalo Calves: A Potential Threat for Animal to Human Zoonotic Transmission
Prasad Minakshi1*, Koushlesh Ranjan2, Neelam Pandey1, Swati Dahiya3, Sandip Khurana4, Neha Bhardwaj5, Sanjeev Bhardwaj5, Gaya Prasad6
1Department of Animal Biotechnology, LLR University of Veterinary and Animal Sciences, Hisar, Haryana, 125004; 2Department of Veterinary Physiology and Biochemistry, Sardar Vallabhbhai Patel University of Agriculture and Technology, Meerut, Uttar Pradesh, 250110; 3Department of Veterinary Microbiology, LLR University of Veterinary and Animal Sciences, Hisar, Haryana, 125004; 4Department National Research centre on Equines, Sirsa road, Hisar, Haryana, 125001; 5Agrionics, CSIR-Central Scientific Instruments Organisation, Chandigarh-160030; 6Indian Council of Agricultural Research (ICAR), New Delhi, India.
Abstract | Gastroenteritis among young dairy calves is predominantly caused by group A rotaviruses (RVA), which leads to calf mortality and significant economic losses to dairy farmers. Huge genetic and antigenic diversity exists amongst different RVA isolates due to segmented nature of the dsRNA genome and diverse host range. The RNA-PAGE analysis of 11 diarrheic fecal samples of buffalo calves from organized dairy farm in Hisar region showed presence of RVA. The VP7 (G type) and VP4 (P type) gene based genotyping of all the sample was carried out by semi nested multiplex RT-PCR. All the samples yielded expected product of 864 bp for VP4 and 1013 bp for VP7 genes, respectively, using generic primers. Nine of 11 (81.81%) samples were categorized as P[11] exhibiting 332 bp product whilst two samples remained un-typed by semi-nested multiplex PCR using cocktail of genotype specific primers for P[1], P[5] and P[11] targeting VP4 gene. The G genotyping of VP7 genes yielded 644 bp products in 8 (72.72%) and were categorized as G10. The remaining 3 (27.27%) samples yielded 288 bp products specific for G6 genotypes. The P genotype for two of the G6 genotypes remained unknown by semi nested multiplex RT-PCR. The 8 (72.72%) isolates with G10 genotype were classified to bear G10 P[11], where, as only one G6 genotyped sample was determined to have G6 P[11] genotype combination. Recently, increasing incidences of involvement of animal rotavirus with G10 specificity in human’s gastroenteritis poses a serious threat of zoonotic transmission of this virus. Hence presence of G10 type in humans is of great concern and widespread surveillance for animal and human rotaviruses is needed to determine the diversity of rotaviruses circulating in various parts of India before adopting any immunization programs.
Keywords | Buffalo rotavirus, Genotype G6, G10, P genotype, RT-PCR, Zoonotic potential
Editor | Varsha Sharma, Deptt. of Vety. Microbiology, College of Vety. Sc.& A.H., South Civil Lines, Jabalpur (M.P), India.
Special Issue | 1(2015) “Biotechnological and molecular approaches for diagnosis, prevention and control of infectious diseases of animals”
Received | January 26, 2015; Revised | March 22, 2015; Accepted | March 23, 2015; Published | April 05, 2015
*Correspondence | Minakshi Prasad, Lala lajpat Rai University of Vetrinary and Animal Sciences, Hisar, Haryana, India; Email: minakshi.abt@gmail.com
Citation | Minakshi P, Ranjan K, Pandey N, Dahiya S, Khurana S, Bhardwaj N, Bhardwaj S, Prasad G (2015). Predominance of G10 genotype of rotavirus in diarrheic buffalo calves: a potential threat for animal to human zoonotic transmission. Adv. Anim. Vet. Sci. 3(1s): 16-21.
DOI | http://dx.doi.org/10.14737/journal.aavs/2015/3.1s.16.21
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2015 Minakshi et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Group A rotaviruses are important enteric pathogens of humans and animals accounting for estimated 611,000 deaths in children each year in developing countries (Ramani and Kang 2007). Rotavirus-associated diarrhea is a major problem in livestock animals including neonatal calves and piglets. Bovine rotaviruses (BoRVs) are responsible for significant economic losses to dairy industry. Buffalo production contributes significantly to the economy of rural farmers in India. Calf mortality due to gastroenteritis is major concern for farmers and animal scientists (Rathore, 1998). The mortality due to Rota viral infection in buffalo calves may reach up to 25% (Fagiolo et al., 2005). Rotaviruses belong to genus Rotavirus and family Reoviridae. The members of this family are characterized by segmented genome which is composed of 11 double stranded RNA (dsRNA) segments surrounded by three protein shells i.e. core, inner capsid and outer capsid. The inner capsid is composed of VP6 protein, encoded by gene segment 6. VP6 bears group specific and subgroup antigenic specificities. Group A rotaviruses of the eight groups of rotaviruses (A-H) have been found to be the most common agents of diarrhea in animals and human beings. The outer capsid consists of two main proteins VP4, a minor component, encoded by gene segment 4 and VP7, a glycoprotein, encoded by genome segments 7, 8 or 9 depending upon the strain. Genotypes of the virus have been defined on the basis of sequences of gene segments encoding VP7 and VP4 proteins. So far 27 G types (VP7 genotypes) have been reported of which G6, G8, G10 belong specifically to bovine origin whereas G1-G4 are of human origin. Of the 37 P (VP4 genotypes) types P1, P5 and P11 belong to bovines whereas P4, P6 and P8 have been identified from humans (Park et al., 2006; Estes and Kapikian 2007; Khamrin et al., 2007; Matthijnssens et al., 2011; Trojnar et al., 2013). Therefore, it is of utmost importance to detect circulating rotaviruses in field and determine their G and P specificities so that a suitable predominant strain could be selected for development of new sensitive diagnostics and efficacious vaccine against rotaviruses.
Many case reports have suggested infection of humans by animal rotaviruses. Comparison of genetic sequences of human and animal rotaviruses often reveals close identity. Presence of several uncommon genotypes among human population similar to that detected in domestic animals suggests their potential threat for zoonotic transmission. This happens within farming communities, especially rural areas where animals and humans are living in very close vicinity. Though this exposure may not result in high levels of infection, but some viral infection continues to be transmitted from animals to humans.
Keeping the above perspective in view, G and P genotyping of buffalo group A rotavirus by RT-PCR has been reported directly in fecal samples of diarrheic buffalo calves.
Materials and methods
Samples Origin
A total of 103 fecal samples from diarrheic calves of 0 to 1 month of age were collected from organized dairy farms of Hisar district of Haryana state during 2007-2008. A 10% suspension (w/v) of faecal samples was made in PBS buffer for viral dsRNA extraction.
Extraction of Viral Nucleic Acid
Viral RNA was extracted by GIT lysis method as described previously (Chomoczynski and Sacchi, 1987; Minakshi et al., 2005). Briefly, to the 500µl of diluted fecal samples, added equal volume of GIT lysis buffer and mixed and kept on ice for 15 min followed by addition of equal volume of phenol: Chloroform: Isoamyl alcohol (PCI) mix (25:24:1) and centrifuged at 10,000 g (REMI cooling centrifuge BL C24) for 10 min at 4oC. The resultant aqueous phase was extracted with equal volume of CI mixture (24:1). The final aqueous phase was kept at -20oC overnight after adding 0.1 volume of 3M sodium acetate, pH 5.2 and equal volume of isopropanol. The dsRNA was pelleted by centrifugation at 10,000g (Remi, India) and dissolved in 20µl of nuclease free water (NFW) and stored at -20oC till further use.
RNA-Polyacrylamide Gel Electrophoresis (RNA-PAGE)
The viral nucleic acid of all the samples were subjected to 8% polyacrylamide gel electrophoresis followed by silver staining to visualize the presence of rotaviral nucleic acid (Svensson et al., 1986).
Amplification of VP4 and VP7 Gene Segments by RT-PCR
The cDNA was synthesized for both VP4 and VP7 genes of viral dsRNA as described earlier (Minakshi et al., 2001; Minakshi and Pandey, 2002) by reverse transcription using MMLV reverse transcriptase (Stratagene, USA) enzyme in 25µl reaction volume. The amplification of cDNA was carried out using VP4 and VP7 gene specific published oligonucleotide primers (Isegawa et al. 1993). PCR reactions were carried out in 25µl volumes containing 2µl cDNA, 1.5µl DMSO and 25pmol of each primer. The mixture was heat denatured at 100oC for 5 minUTE, snap chilled on ice and added 5 µl of PCR mixture consisting of 2.5 µl 10x PCR buffer, 0.5 µl 100 mM dNTPs, 1.5 µl 25mM MgCl2, 0.5 µl Taq polymerase (5 U/µl) (MBI, Fermentas). The NFW was added to make final volume 25 µl. The PCR reactions were performed on thermal-cycler (Biorad, USA). The cycling conditions consisted of one cycle of initial denaturation at 94ºC for 5 minute, 30 cycles of amplification consisted of denaturation at 94ºC for 1 minute, annealing at 45ºC for 2 minute and elongation at 72ºC for 3 minute. This was followed by final extension at 72ºC for 7 minute. The products were visualized in 1% agarose gel electrophoresis under UV transilluminator (Biovis, USA).
P and G Genotyping of Amplified VP4 (P) and VP7 (G) Genes
The VP4 and VP7 gene specific PCR products were diluted to 1:100 and used as template for semi-nested multiplex PCR for P (VP4) and G (VP7) genotyping. The reaction mixture was prepared for 50 µl containing 2 µl of template, 10 µM of each Bov4Com5 (plus sense) primer, cocktail of typing primers (P[1], P[5], P[11]) for VP4 and Bov9Com5 (plus sense) primer, typing primers (G6 and G10) for VP7 gene, 200 µM each of dNTP, 1X PCR buffer, 1.5 mM MgCl2 and 2.5 units of Taq polymerase (MBI Fermentas, USA). Amplification was carried out by initial denaturation at 94ºC for 2 minute, 25 cycles of amplification consisting denaturation at 94ºC for 1 minute, annealing at 50ºC for 2 minute and elongation at 72ºC for 3 minute, followed by final extension at 72ºC for 7 minute. The nested PCR products were visualized in 1% agarose gel under UV transilluminator (Biovis, USA) and genotype was assigned according to the size of PCR products.
Results
RNA-PAGE
A total of 11 samples showed eleven segmented genome and were found positive for rotavirus in RNA-PAGE analysis. The nucleic acid migration pattern of all the buffalo calf samples showed 4:2:3:2 migration patterns, typical of long electropherotype of bovine group A rotaviruses (data not shown).
RT-PCR for VP4 and VP7 Genes
The RT-PCR of viral cDNA of all the eleven samples showed a PCR amplicons of 1013 bp and 864 bp specific to VP7 and VP4 genes respectively as evidenced by 1% agarose gel electrophoresis (Figure 1A and 1B).
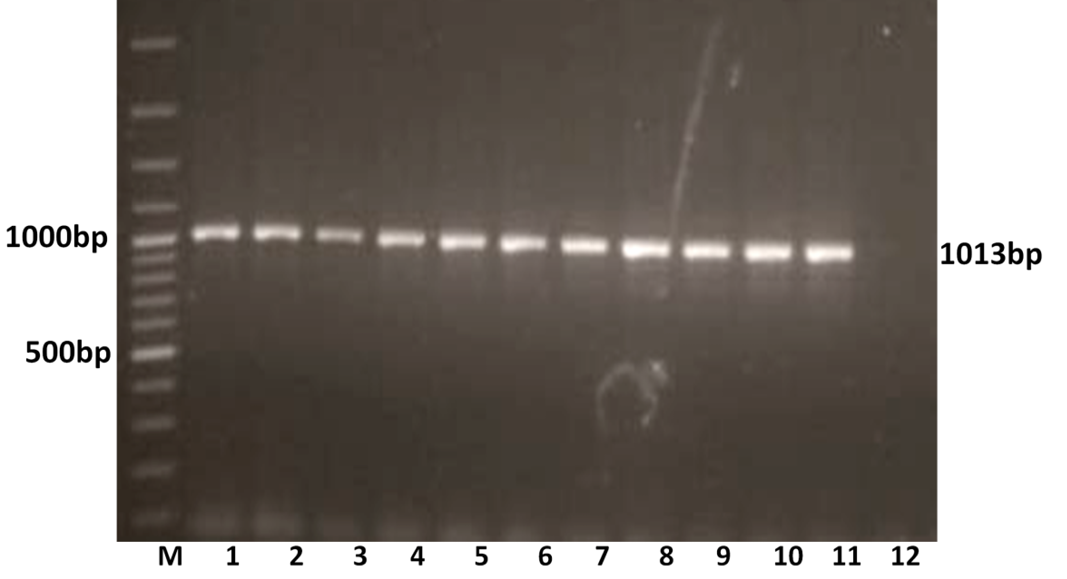
Figure 1A: Detection of amplification of cDNA of VP7 genes of buffalo rotavirus in l% agarose gel electrophoresis of buffalo rotavirus Isolates by RT- PCR
Lanes: M- 100 bp DNA Ladder, 1-11: BoRVA VP7 gene products, 12: no template control.
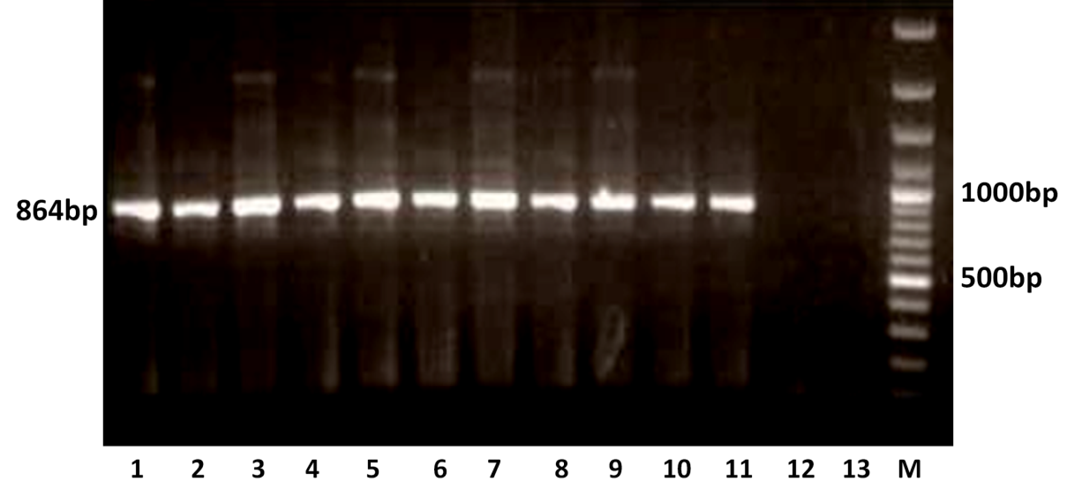
Figure 1B: Detection of amplification of cDNA of VP4 genes of buffalo rotavirus in l % agarose gel electrophoresis of buffalo rotavirus Isolates by RT- PCR
Lanes: M- 100 bp DNA Ladder, 1-11: BoRVA VP4 gene products, 12-13: no template control.
P Genotyping of BoRVA by Semi-nested Multiplex PCR
The VP4 gene specific PCR products of all the samples were further subjected to semi-nested PCR to ascertain the genotype using type specific primers for P[1], P[5] and P[11] genotypes. Out of 11 samples amplified by RT-PCR, 9 samples were genotyped as P[11] as evidenced by a single 332 bp of PCR amplicon (expected product of P[11] genotype) in 1% agarose gel (Figure 2). However, other two samples remain untyped by semi-nested PCR.
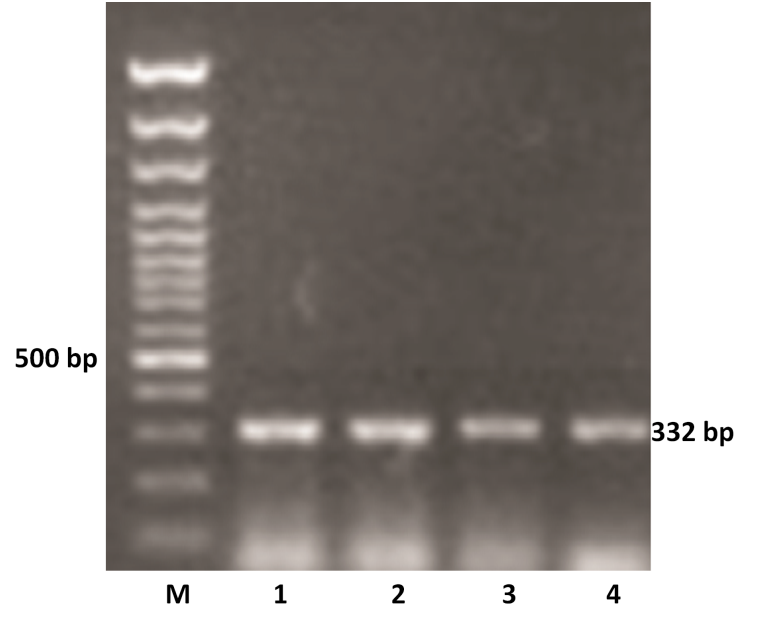
Figure 2: P genotyping of the buffalo rotavirus using VP4 gene type specific primers by semi-nested multiplex PCR
Lanes: M- 100 bp DNA Ladder, 1-4: BoRVA P[11] genotype specific PCR products.
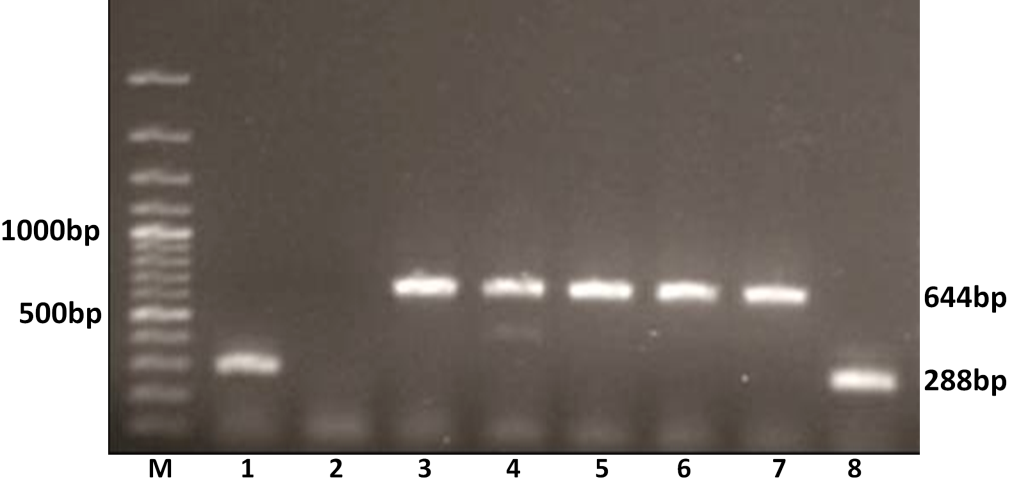
Figure 3: G genotyping of the buffalo rotavirus using VP7 genotypes specific primers by semi-nested multiplex PCR
Lanes: M- 100 bp DNA Ladder; Lanes: 1 and 8 BoRVA G6 genotype specific products; 2: no template control; 3-7: BoRVA G10 genotype specific products.
G Genotyping of BoRVA by Semi-nested Multiplex PCR
The G typing using G6 and G10 primers revealed that out of 11 amplified samples, 8 (72.72 %) were yielded PCR amplicon of 644 bp, an expected size of G10 genotype. The remaining 3 (27.27%) samples showed G6 genotype specific an expected amplicon of 288 bp products in 1% agarose gel electrophoresis (Figure 3). Thus 8 samples were genotyped as G10 and 3 as G6 genotype.
Discussion
The vast diversity of rotaviruses circulating in India highlights the need for uniform and widespread surveillance. The buffalo calves diarrheal samples were preliminary examined by RNA-PAGE analysis and rotavirus detected in 11 samples as indicated by 4:2:3:2 migration pattern of 11 dsRNA segments of group A rotavirus. All the RNA-PAGE positive samples yielded a specific product of 1013bp for VP7 and 864 bp for VP4 genes in RT-PCR. The VP4 gene amplified PCR products subsequently subjected to semi-nested multiplex PCR to ascertain the genotype using type specific primers for P[1], P[5] and P[11]; 9 samples (81.81%) were categorized as P[11] and 2 samples remained un-typed. Similarly, the VP7 gene based genotyping revealed 8 samples (72.73%) as G10 and 3 samples (27.27%) as G6 genotype. The 8 samples (72.72%) of G10 genotype were assigned G10 P[11] genotype whereas only one sample with G6 genotype was assigned G6P[11] genotype. However, P genotype for two G6 genotypes remained unknown by RT-PCR genotyping. The un-typed isolates might belong to some unusual genotypes of rotaviruses.
The results are in conformity with the finding obtained in various studies carried out in various parts of India such as Haryana, Gujarat, Kashmir, Mumbai, Tamil Nadu and Uttar Pradesh where G10 detected as predominant genotype followed by G6 with G10P [11] predominant combination followed by G6P [1] and G6P [11] (Beg et al., 2010; Gulati et al., 1995, 1999; Mondal et al., 2012; Minakshi et al., 2005; Mittal et al., 1986; Saravanan et al., 2006; Singh and Jhala, 2011; Suresh et al., 2012; Wani et al. 2004).
Group A rotaviruses are important enteric pathogens of humans and animals accounting for high mortality in humans and neonates of animals specifically in developing countries (Ramani and kang, 2007). Rotavirus-associated diarrhea is a major problem in livestock animals including neonatal calves and piglets. Many unusual genotypes of animal and human rotaviruses have been revealed (Manuja et al., 2008; Esona, et al. 2011). The G10 has also been reported as mixed infection along with other G genotypes of human and animal origin such as G3G10 or G3G8 genotypes in bovine samples (Malik et al., 2012).
Incidences of cross-species transmission of rotaviruses have been increased reportedly in recent years and G10 rotaviruses, which are usually found in buffalo, cattle, have also been reported in human neonatal infections. Genetic heterogeneity has been reported in all the eleven double stranded RNA segments of rotaviruses of human and animal origin through whole genome sequencing (Ramani et al., 2009). The data has provided evidence for frequent evolution of human and animal rotaviruses resulting from interspecies transmission and adaptation of rotaviruses.
In India initial report on detection of unusual G10P[14] genotype from human rotavirus was reported from West Bengal (Ghosh et al., 2007) and subsequently in Kolkata, where detection of human G10 rotavirus strains was done from untypable sample from eastern India (Mukherjee et al., 2012). In Tamil Nadu and Pune G10 P[11] genotype was reported from rotavirus infected children (Kelkar and Ayachit, 2000; Paul et al., 2014). Similarly, the studies in Vellore region showed prevalence of G10 in human, bovine and caprine indicating the interspecies transmission by G10 genotype (Rajendran and Kang, 2014). Ramani and coworkers in 2009 revealed genetic reassortant of vp4 [11] and vp7 (G10) gene segments of a human neonate strain (N155) infected with rotavirus were of animal origin G10 P[11] and identified other genes of human and animal rotavirus origin on further characterization. In another finding, genotype G10 was detected in human in Vietnam. That human rotavirus was characterized as G10 P[8] consisting of G10 from bovines and P[8] of human origin (Matsushima et al., 2012). Another case report from rotavirus infected humans indicated a genotype G10 P[14] of human rotavirus isolated from 5 children and 1 adult with acute gastroenteritis in Australia. Full genome sequence analysis identified an artiodactyl-like (bovine, ovine, and camelid) G10-P[14]-I2-R2-C2-M2-A11-N2-T6-E2-H3 genome constellation (Cowley et al., 2013).
Though the zoonotic potential of G10P[11] genotype of bovine strain was evidenced by its transmission from cattle to humans and vice-versa (Iturriza-Gomara et al. 2004; Saravanan et al., 2006), the reports on zoonotic potential of buffalo G10 rotavirus is lacking in literature. Buffalo is of high economic value in India and very expensive milch animal, treated as family members in rural areas. Their presence in close vicinity to human may pose potential zoonotic threat to human. In this study, we reported G6 P[11] and G10P[11] genotypes from buffalo calves. It may be an important contributor of human rotavirus genomic diversity. These finding suggests animal to human transmission and strengthens the need to continue rotavirus strain surveillance.
Acknowledgements
The study was funded by ICAR, New Delhi, The authors are thankful to Department of Animal Biotechnology, LLR University of Veterinary and Animal Sciences, Hisar for providing infrastructural facility.
References





