Advances in Animal and Veterinary Sciences
Research Article
Prevalence of Aflatoxin M1 and Molecular Studies on Some Food Born Fungi Isolated from Milk and Dairy Products
Ashraf Awad Abd El-Tawab1, Fatma Ibrahim El-Hofy1, Eman Mahmoud EL-Diasty2, Ebtehal Ahmed Abo-Hamdah2*, Manar El-Hayat1
1Bacteriology, Immunology and Mycology Department, Faculty of Veterinary Medicine, Benha University, Egypt; 2Animal Health Research Institute, Dokki, Giza (ARC), Egypt.
Abstract | Fungal contamination of milk and dairy products is the main cause of spoilage of these products and mycotoxicosis in humans. The aim of this study was to investigate the fungal contamination of milk and some dairy products. One hundred and twenty samples of milk and dairy products (20 of each processed cheese, Talaga cheese, powdered milk, UHT milk, pasteurized milk and raw milk) were collected from different markets at Giza Governorate, Egypt and then submitted for mycological, molecular examination and detection of Aflatoxin M1residues (AFM1). The mycological examination revealed that the fungal contamination was not detected in Ultra high temperature (UHT) milk samples, whereas yeast contamination was detected only in Talaga cheese and raw milk with incidence rate of 50 % and 80%, respectively. In addition, the prevalence of moulds in processed cheese, Talaga cheese, powdered milk, pasteurized milk and raw milk was 90%, 30%, 35%, 30% and 80% respectively. Moreover, AFM1 residues were detected in Talaga cheese, powdered milk, UHT milk, pasteurized milk and raw milk with a prevalence of 20%,15%, 10%,20% and 15%, respectively. In this analysis, we have optimized the conditions for the use of real-time PCR (RT-PCR) together with propidium monoazide (PMA), (q PCR-PMA) for discrimination between the live and dead fungal cells in the examined samples. Nine samples of milk and dairy products, which were negative upon culturing, were subjected to ITS SYBR green RT- PCR, with the results showing that six out of the nine tested samples were found to be positive. After words, these six samples were subjected to propidium monoazide (PMA) treatment prior to RT-PCR to demonstrate the viability of the fungal cells. As a result, all of the six positive samples in RT-PCR were positive after PMA treatment, indicating that all positive RT-PCR samples contained viable but non-culturable fungal cells. In conclusion, Propidium monoazide (PMA) treatment accompanied with RT- PCR is a rapid and reliable method for molecular identification of fungal contamination in milk and milk products.
Keywords | Fungi, Milk products, AFM1, RT-PCR, PMA, Egypt
Received | December 22, 2019; Accepted | February 17, 2020; Published | March 05, 2020
*Correspondence | Ebtehal Ahmed Abo-Hamdah, Animal Health Research Institute, Dokki, Giza (ARC); Email: ebtehal.abohamda@gmail.com
Citation | El-Tawab AAA, El-Hofy FI, EL-Diasty EM, Abo-Hamdah EA, El-Hayat M (2020). Prevalence of aflatoxin m1 and molecular studies on some food born fungi isolated from milk and dairy products. Adv. Anim. Vet. Sci. 8(3): 305-311.
DOI | http://dx.doi.org/10.17582/journal.aavs/2020/8.3.305.311
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2020 El-Tawab et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
Many people consume milk and dairy products on a daily basis, especially for the growing infant population, which depend on milk as principle nutrient. So fungal contamination and aflatoxin M1 occurrence in milk and dairy products is serious problem. (Elkak et al., 2012). Also, fungal spoilage is a perilous problem in the dairy products industry, since raw milk and milk products are generally considered an ideal growth medium for many fungal species; as they provide all the important nutrients for their growth (Callon et al., 2007; Gulbe and Valdovska, 2014). Fungal contamination of raw milk may occur during milking, storage and other pre-processing practices and influenced by the animal’s physiological state, breeding condition and the weather (Callon et al., 2007). In addition, it represents an obstacle to increased demand by consumers and manufacturers for natural dairy products, especially with increased fungal resistance to heat treatment and chemical preservatives (Garnier et al., 2017). For milk and dairy products, the identification and characterization of fungi was based on the phenotypic and genotypic characteristics. The fungal count was also used as an index of the dairy product’s storability and sanitary quality (Moubasher et al., 2018). Mycotoxines, especially aflatoxins, are hazardous fungal toxins causing mycotoxicosis in humans and animals that are characterized by immune system disorders, liver cancers, children’s growth disorders and deaths in humans and animals. (Becker-Algeri et al., 2016). The most common mycotoxins produced by fungi are Aflatoxins especially of the genus Aspergillus, particularly A. flavus and A. parasiticus. The feeding of dairy animals on feedstuffs contaminated with aflatoxin B1 (AFB1) lead to this toxin passes into animal’s urine and milk and detection of aflatoxin M1 (AFM1) in milk. Therefore, the evaluation of the AFM1 concentration in milk is the best study for this phenomenon control to give a good level of safety for the human consumption (Sottili et al., 2017). Also, the elimination of fungus-susceptible animal feed ingredients is a need, as it represents the main source of mycotoxins in milk and dairy products (Becker-Algeri et al., 2016). The advent of real-time RT-PCR for identification of fungal contamination and the availability of modern equipment provide significant prospects for the accurate quantification of mRNA species. RT-PCR has several benefits over other PCR based quantification approaches, such as processing of large numbers of samples, easier automation and elimination of post amplification handling. In addition, it has a very large dynamic range of template determination (Heid et al., 1996). Real-time PCR combined with the use of Propidium monoazaid (PMA), was recently introduced. Propidium monoazaid is a binding dye with high affinity reactive DNA used by scientists for viability PCR of fungi. PMA, which enters dead cells but not live cells, was incubated with cell suspensions, exposed to blue wavelength light-emitting diodes (LED) to inactivate the remaining PMA and save the intercalation of PMA with DNA of dead cells. Treated cells with PMA were extracted then the dead and live cells were evaluated with quantitative PCR (QPCR) (Stephen et al., 2008). The present study was aimed to detect Aflatoxin M1 (AFM1) in milk and some dairy products with optimization of conditions for using of real-time PCR together with PMA for discrimination between the live and dead fungal cells present in the examined milk and dairy product samples.
MATERIALS AND METHODS
Collection of samples
A total of 120 milk and dairy product samples (20 of each); processed cheeses, Talaga cheeses, milk powder, UHT milk, pasteurized milk and raw milk) were collected from various groceries and supermarkets in Giza Governorate, Egypt from March 2018 to September 2019. All of these samples were collected and transferred aseptically in the icebox to the laboratory.
Preparation of samples (APHA, 2001)
Prparation of cheese (APHA, 1985): Eleven grams of each prepared cheese sample were removed aseptically and transferred in a sterile homogenizer flask containing 99 ml of sterile (2%) Sodium citrate. The contents were homogenized at 14000 rpm for 2.5 minutes to provide a dilution of 1:10. One ml from cheese homogenate was transferred to separate sterile test tubes containing 9 ml of sterile peptone water (1%) from which ten–fold serial dilutions up to 10-6 were prepared.
Preparation of milk samples (APHA, 2001): Twenty-five grams of each sample were aseptically homogenized in a blender with 225 ml of sterile peptone water to make a dilution of 1:10, from which 10th fold serial dilutions were accomplished up to 106. Concerning milk powder, Eleven grams of each prepared dried milk powder sample were removed aseptically and transferred in a sterile homogenizer flask containing 99 ml of Dipotassium hydrogen phosphate solution (pH 7.5). Thoroughly the contents of the closed sample container mixed by repeatedly shaking and inverting. If the container is too full to allow thorough mixing, the contents transferred to a larger container and mixed. In order to dissolve, swirl slowly to wet the powder, then the bottle was shaked 25 times in about 7 seconds or mixed in a stomacher. Allowed to stand for 5 minutes, shaking occasionally. One ml from Dried milk powder homogenate was transferred to separate sterile test tubes containing 9 ml of peptone water (1%) from which ten–fold serial dilutions up to 10–6
Isolation of fungal species
a) It was done according to (ISO, 2008). Using Dichloran Rose Bengal Chloramphenicol (DRBC) agar supplemented with chloramphenicol (0.05 mg /liter).
b) By sterile pipette, 0.1 ml from each dilution was transferred to a single DRBC agar plate and distributed over the surface of the agar plate by sterile spreader. The inoculated plates were incubated in an inverted position at 25 0C for 5-7 days. Suspected colonies were subsequently picked up from the DRBC agar plates onto the Sabouraud dextrose agar (SDA) slopes (Oxoid CM0041) and incubated at 25°C C for 5-7 days, subsequently sub cultured onto malt extract agar (MEA) (Oxoid CM0059) and Czapek yeast agar (CYA) (Pitt and Hocking, 2009) plates, and incubated at 25°C for one week.
Identification of fungal isolates
Identification of moulds: Mould colonies were identified by examining their Macroscopic and Microscopic features according to Pitt and Hocking (2009).
Identification of yeasts: Suspected yeast colonies were subcultured onto Sabouraud Dextrose agar slopes and incubated at 250C for48 hrs. Then the identification of yeasts was performed according to Lodder and Krger-Van Rij (1970).
Detection of aflatoxin M1 residues in milk and milk products by fluorometer (VICAM manual 2014)
Afla test Fluorometer procedures for milk and cheese samples were done according to AOAC (1995). (0-2.0 PPB) and (0-50 PPB), respectively. The aflatoxin M1 concentration was measured in a fluorometer (serial No. 0748 USA. Model No. VICAM series 4 and 4EX 1107-103606.
Molecular Identification of Fungal Isolates by RT-PCR 3.6.
Extraction of genomic DNA from fungal isolates: Fungal isolates were submitted for extraction of genomic DNA by using the DNeasy Plant Mini Kit (Qiagen, Catalogue no. 69104).
Molecular identification of fungal isolates by ITS SYBR green RT–PCR (Mirhendi et al., 2007): Fungal isolates were submitted for QRT-PCR using the forward primer (ITS.1) TCC GTA GGT GAA CCT GCG G and the reverse primer (ITS. 4) TCC TCC GCT TAT TGA TAT GC targeting the ITS gene of fungal spp. (Mirhendi et al., 2007). The reaction was carried out on Strata gene Mx3005P QPCR system (Agilent Technologies) in a total reaction volume of 25 µl containing 12.5µl 2X Quantitect SYBR green PCR Master Mix (Qiagen, Cat. No. 204141), 1.0 µl of magnesium chloride sol. 25mM, 0.5 µl. forward primer (20 pmol), 0.5 µl reverse primer (20 pmol), 6.5µl nuclease free molecular biology grade Water and 5µl test DNA. The cycling conditions for SYBR green real time PCR program was adjusted as the following: Primary denaturation thermo start activation step 95°C for 5 min 1 cycle, and Secondary denaturation 94°C for 15 sec 40 cycle, annealing 56°C for 30 sec and extension 72°C for 30 sec. At dissociation curve the secondary denaturation 95°C for 30 sec. 1 cycle, annealing 56°C for 30 sec (optics on till final denaturation) final denaturation 95°C for 30 sec. Interpretation of results was made and read after the end of each extension step.
Identification of fungal Isolates by propidium iodide staining with RT-PCR(PMA-RT.PCR): It was performed according to Taskin et al. (2011) with the following modification:
A10 mM PMA stock solution was prepared by dissolving Propidium iodide (Sigma-Aldrich, P4170) in 20% dimethyl sulfoxide (DMSO; Sigma-Aldrich, St. Louis, MO) and stored at -20 °C. Then PMA stock solution was transferred into 500 µl culture mixtures at a final concentration of 100 µM followed by stored at −20°C in the dark for10 min before used. After wards, samples were exposed to a 500-Whalogen light source (Deng et al., 2016) for 5 minutes at a distance of 15 to 20 cm from the light source with the tubes placed on ice during the light exposure to avoid excessive heating. Then DNA was extracted according to QIA amp DNA mini kit instructions and real-time PCR was performed with the PMA treated samples, then the CT values were calculated and prevalence of viable but non -culturable fungal cells were determined.
Results
One hundred and twenty samples of milk and milk products (20 of each, processed cheese, Talaga cheese, powdered milk, UHT milk, pasteurized milk and raw milk) were collected from supermarkets in Giza Governorate, Egypt and submitted for mycological, molecular examination and Aflatoxin M1 detection. Mycological examination revealed that UHT milk samples were negative for both mould and yeast contamination while yeast contamination was detected only in Talaga cheese and raw milk with prevalence of 50 % and 80%, respectively. In addition, the prevalence of the moulds in processed cheese, Talaga cheese, powdered milk, pasteurized milk and raw milk were 90%, 30%, 35%, 30% and 80%, respectively (Table 1). Moreover, the results showed that the most prevalent isolated genera in processed cheese were Aspergillus spp. with prevalence of 36(62%), followed by Penicillium spp. 10(17.2%), Eupenicillium spp. 6 (10.3%), Fusariumspp 4(7%) and Phoma spp. 2(3.5%). The most prevalent genera isolated from Talaga cheese were Aspergillus spp. with prevalence of 10 (83.3%), followed by Penicillium spp. 2(16.7%). The most prevalent genera in powdered milk were Aspergillus spp. 4(50%), Penicillium spp. 2(25%) and Alternaria spp. 2(25%). While in pasteurized milk the most prevalent genera were Aspergillus spp. with prevalence of 2(33.3%), Penicillium spp. 2(33.3%). and Epicoceum spp. 2(33.3%). The most prevalent genera in raw milk was Penicillium spp. with prevalence of 16(44.5%), followed by Aspergillus spp. 14(38.9%) and Geotrichum spp. 6 (16.6%). additionally, the foremost prevailing yeast species isolated from Talaga cheese samples were Debaryomyceshansenii 14(53.8%) followed by Pichia anomala 8 (30.8%), Pichiamembrana faciens 2(7.7%) and Sacharomyces cervisioe 2(7.7). Also, the most prevalent yeast species in the examined raw milk samples were C. tropicalis, C. pseudotropicalis, Sacharomyces cervisioe and Torulopsis with an incidence of 2(14.3%) of each as well as Rhodotorula spp. 6(42.8%).
Table 1: The prevalence of moulds and yeasts isolated from the examined samples.
| Samples | No. of examined samples | Mould | Yeast | ||||
| +ve | % | Mean ± SE cfu/g | +ve | % | Mean ± SE cfu/g | ||
| Processed cheese | 20 | 18 | 90 |
1.3 × 103 ± 1.8× 10² |
0 | 0 | 0 |
| Talaga cheese | 20 | 6 | 30 |
6.7 × 103 ± 2.6× 103 |
10 | 50 | 5 × 10 ± 1.3× 10 |
| Powdered milk | 20 | 7 | 35 | 1.2 × 10² ± 3.7× 10 | 0 | 0 | 0 |
| UHT milk | 20 | 0 | 0 | 0 | 0 | 0 | 0 |
| Pasteurized milk | 20 | 6 | 30 | 2.1 × 10² ± 9.9× 10 | 0 | 0 | 0 |
| Raw milk | 20 | 16 | 80 |
5.9 × 103 ± 1.3× 103 |
16 | 80 |
7.6 × 102 ± 2.4× 102 |
Regarding aflatoxin M1, it could be detected with prevalence of 20%, 15%, 10%, 20% and 15%, respectively (Table 2). The results were the mean of triplicates + standard error (SE) as in (Table 2).
Table 2: Aflatoxin M1 (AFM1) concentrations (ppb) in different examined samples.
| Type of samples | No. of examined samples | % | Min. | Max. | Mean of level ± SE |
| Processed cheese | 20 | 0 | 0 | 0 | 0 |
| Talaga cheese | 20 | 20 | 3 | 11 | 6.2 ± 1.8 |
| Powder milk | 20 | 15 | 0.52 | 0.9 | 0.73 ± 0.11 |
| UHT milk | 20 | 10 | 0.61 | 0.78 | - |
| Pasteurized milk | 20 | 20 | 0.44 | 1 | 0.59 ± 0.14 |
| Raw milk | 20 | 15 | 0.66 | 0.69 | 0.67 ± 0.01 |
Table 3: Results of ITS SYBR green RT-PCR and PMA treatment of nine negative examined samples.
| Sample | Type of sample |
ITS SYBR green RT-PCR |
RT-PCR After PMA treatment |
| 1 and2 | Talaga cheese | + | + |
| 3 | Powder milk | + | + |
| 4 | Powder milk | - | - |
| 5and6 | UHT milk | - | - |
| 7 | Raw milk | + | + |
| 8and9 | Pasteurized milk | + | + |
In the present study, we have got optimized the conditions for used RT-PCR along with PMA (q PCR-PMA) for discrimination between the live and dead fungal cells present in milk and dairy product samples. Nine milk and dairy product samples, that were negative upon culturing, were subjected to ITS SYBR green real time PCR. Out of the nine samples, six were found to be positive for fungal contamination by ITS SYBR green real time PCR. However, there are two possibilities to those positive samples, either they contain dead cells therefore failed to grow on the culture or it may be viable but non-culturable cells (VBNC). So, the six positive samples in real time PCR were subjected to propidium monoazide (PMA) treatment, prior to RT- PCR. The results disclosed that each one of the six positive samples in RT-PCR were positive after PMA treatment, that indicated that all the positive samples in RT-PCR were containing VBNC fungi. (Table 3, Figures 1, 2 and 3).
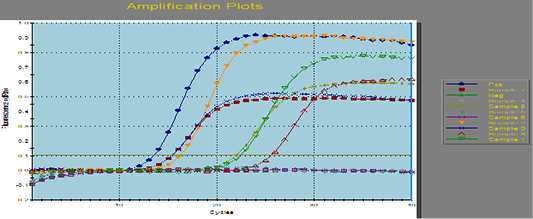
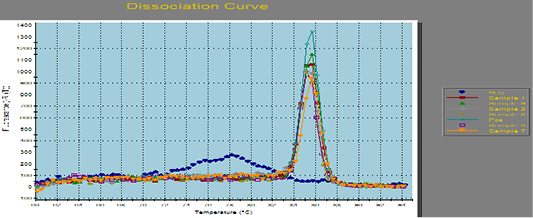
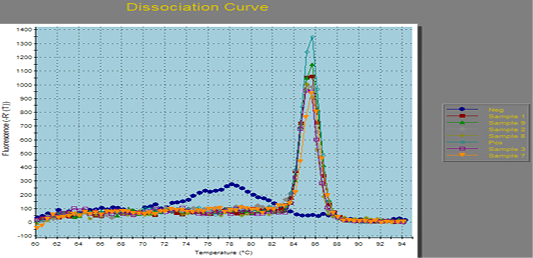
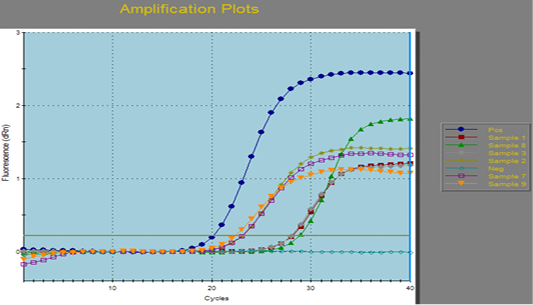
Figure 1: Amplification curves of SYBR green RT-PCR for fungal ITS gene. RT-PCR results for fungal ITS gene showing positive amplification of ITS sequence gene solely within the tested sample. Positive amplification and negative amplification of the various ITS fungal genes were reported.
Aspergillus fumigates field sample were portrayed the Positive and negative controls and by using PCR for the related fungal genes (ITS) it antecedently confirmed to be positive or negative within the reference laboratory for veterinary internal quality control on poultry production, Animal health research institute.
DISCUSSION
Fungal contamination of raw milk and milk products may happens throughout milking, storage and alternative pre-processing activities and influenced by the animal’s physiological condition, breeding condition and the weather (Callon et al., 2007). The results revealed that fungal contamination was not detected in all examined UHT milk samples. The prevalence of the moulds in processed cheese, Talaga cheese, powdered milk, pasteurized milk and raw milk were 90%, 30%, 35%, 30% and 80%, respectively with mean mould counts, 1.3 × 103 ± 1.8× 10² cfu/g, 6.7 × 103 ± 2.6× 103 cfu/g, 1.2 × 10² ± 3.7× 10 cfu/g, 2.1 × 10² ± 9.9× 10 cfu/L, 5.9 × 103 ± 1.3× 103 cfu/L, respectively (Table 1).
The obtained results were nearly similar to Sabreen and Zaky (2001), Amer (2002). While EL-Bagory et al. (2014) and Younis et al. (2016), were obtained higher results.
High fungal prevalence up to 15 species were detected in the samples of cow milks, also a maximum of 4 or 6 different species were isolated from goat and sheep milks, respectively. (Delavenne et al., 2011). In the present study, the most common isolated mould species from the examined samples were Aspergillus spp., Penicillium spp., Eupenicillium spp., Fusarium spp., Phoma spp. and Geotrichum spp. These results nearly similar to the results obtained by Hassan (2010), Nolwenn et al. (2014) and Silva et al. (2015) who reported that the most isolated fungi producing mycotoxins in cheese and milk were Aspergillus spp., Penicillium spp., Geotrichum spp. and Fusarium spp. Moreover, the present results were in agreement with those obtained by Guirguis (2015) who reported that fungal contamination was not detected in infant formulas and UHT milk.
In our study, processed cheese, powdered milk, UHT milk and Pasteurized milk samples showed negative results for the yeast contamination. While Talaga cheese and raw milk were positive for yeast contamination with prevalence of 50% and 80%, respectively (Table 1). The most predominant yeast species isolated from Talaga cheese and raw milk were Debaryomyces hansenii, Pichia anomala, Sacharomycescervisioe, C. tropicalis, C. pseudotropicalis, Torulopsis and Rhodotorula. These results in coordinated with the EOSQ (2005) which stated that processed cheese should be free from any visible fungal contamination.
The prevalence of yeasts in the examined Talaga cheese samples was ranged from 60- 66% (El-Shazly, 2007; Sayed et al., 2011).
The aflatoxines especially AFM1 which produced by certain fungal species leads to spoilage of the dairy products and regarded as a major problem to human and animal health, as it responsible for different adverse health effects and economic loss (Garnier et al., 2017). Also, Egyptian regulation (Egyptian Regulation Standards, 1990) for AFM1 in milk recommended that milk and milk products must be free from AFM1. In this study, AFM1were detected in the examined Talaga cheese, powdered milk, UHT milk, Pasteurized milk and raw milk samples with an incidence of 20%, 15%, 10%, 20% and 15%, respectively (Table 2). These results were nearly in coordinating with those reported by Awad et al. (2014). While higher results were obtained by El-Seadawy et al. (2000) who reported that the prevalence of AFM1 in 50 examined Talaga cheese was 36 samples (18%) which were positive. Culture methods only allow for counting viable cells that are capable of forming colonies on nutrient media, without detecting dead cells, viable but non-culturable bacterial cells (VBNC) and those that require special growth conditions (Cerca et al., 2011). While RT-PCR detects all cells in a sample, including DNA of the dead cells (Pathak et al., 2012). Recently, Real-time PCR (qPCR) combined with the use of propidium monoazaid (PMA), which is a binding dye with high affinity reactive DNA used, by scientists for viability PCR of fungi. PMA enters dead cells but not live cells, was incubated with cell suspensions, exposed to blue wavelength light-emitting diodes (LED) to inactivate the remaining PMA and protect intercalation of PMA with DNA of dead cells. Treated cells with PMA were extracted and the dead and live cells evaluated with real time PCR (RT-PCR) (Stephen et al., 2008). In addition, the parameters of PMA treatment were affected by the cell concentrations, species and RT-PCR kit used for detection. Higher concentrations of PMA inhibit the PCR for live cells and give higher CT values in dead cells (Younis et al., 2016).
In the present study nine milk and dairy samples were negative upon culturing but whene subjected to ITS SYBR green real time PCR, six of them were positive with real time PCR, this indicated presence of fungi in those six samples. However, there are two possibilities to those positive samples, either they contain dead cells so did not grow on the culture or may be viable but non- culturable cells (VBNC). So, the six positive samples in RT-PCR were subjected to propidium mono azide (PMA), prior to RT- PCR., as a result all of the six positive samples in RT-PCR was positive after PMA treatment and this indicated that all positive samples in RT-PCR containing VBNC fungi.
These results agreed with those obtained by Nguyen et al. (2017) who reported that the quantitative PCR method was compared to traditional plate counting on DRBC agar and currently commercial methods in 38 examined dairy products samples (fluid milk, yogurt cottage cheese, sour cream, and cheese).
The results reported that 16 of the 38 samples exhibited amplification by using the qPCR method. Also, Andorrà et al. (2010) found that the under estimation caused by presence of viable but non-countable microorganisms or variable growing rates of different microorganisms in culture media. These results revealed that the presence of excess dead cells did not interfere with the quantification of live cells with the PMA dye and quantitative PCR. While Nguyen et al. (2017) used PMA to bind DNA present from dead cells for q PCR. The results showed that using PMA did not help discern differences in live and dead cells. Also, a potential binding agent as methylene blue was used, however at the same cell concentration there is no differences in Ct values were detected for live and dead cells.
CONCLUSION
Presence of fungal contamination and AFM1 in milk and dairy products with high levels causes economic losses and an important public health hazards. So, we need to control this contamination and strict hygienic measures. In addition, we need to the development of quantification methods and rapid detection into milk, milk products and other foodstuffs by PMA-QPCR which more accurate and sensitive to protect the animal and human health.
Authors Contribution
All authors contributed equally.
Conflict of interest
Herein we confirm that there are no conflict of interest of any type.
REFERENCES