Advances in Animal and Veterinary Sciences
Research Article
Mitochondrial DNA D-Loop Variation among Four Sheep Breeds and Evolutionary Relationships with Other Taxa: Forensic Implication
Yasmina M. Abd-Elhakim1*, Amir H. Abd El-Fattah2, Walaa M. Elhady1, Mayada R. Farag1
1Department of Forensic Medicine and Toxicology, Faculty of Veterinary Medicine, Zagazig University, 44511 Zagazig, Egypt; 2Animal Wealth Development Department, Faculty of Veterinary Medicine, Zagazig University, 44511 Zagazig, Egypt.
Abstract | Animal identification is a very vital issue in both genetic and forensic fields. Mitochondrial DNA (mtDNA) analysis has been often used to explore haplotype variety among and within animal species. The mtDNA displacement-loop (D-loop) region is highly variable than other DNA regions and it is regularly utilized to investigate the phylogeny of highly correlated breeds within species. In this investigation, the mtDNA D-loop sequence was analyzed to evaluate the genetic diversity and origin of four Egyptian sheep breeds (Ossimi, Baladi, Rahmani, and Barki). Additionally, phylogenetic analysis was performed through alignment of the obtained sequences with those available in the GenBank database (goat, pig, buffalo, horse, donkey, and dog). Sequence analysis of the mtDNA D-loop revealed a clustering of the Ossimi and Baladi sheep breeds (haplotype A), while the Rahmani and Barki sheep were observed to cluster (haplotype B). The pairwise differences among the four Egyptian sheep breeds and other sequences indicated a high genetic distance from the dog and the maximum identity was clearly established with goat and pig. In conclusion, this information can help to authenticate sheep species from other taxa and identify the origin of meat of commonly consumed species (cattle, buffalo, goat, and pig) and commonly banned species (dog, horse, and donkey) for food control and forensic purposes.
Keywords | Mitochondrial control region, Phylogenetic analysis, Sheep, Meat adulteration, Forensic identification.
Received | June 25, 2019; Accepted | August 19, 2019; Published | December 08, 2019
*Correspondence | Yasmina M. Abd- Elhakim, Department of Forensic Medicine and Toxicology, Faculty of Veterinary Medicine, Zagazig University, 44511 Zagazig, Egypt; Email: yasmina_forensic@hotmail.com
Citation | Abd-Elhakim YM, Abd El-Fattah AH, M. Elhady W, Farag MR (2020). Mitochondrial dna d-loop variation among four sheep breeds and evolutionary relationships with other taxa: forensic implication. Adv. Anim. Vet. Sci. 8(1): 18-24.
DOI | http://dx.doi.org/10.17582/journal.aavs/2020/8.1.18.24
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2020 Abd-Elhakim et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
The analysis of nonhuman DNA has received increased consideration, particularly in the area of forensic science, ecological investigations, and food control (Druml & Cichna-Markl, 2014). In particular, veterinary and legal science research laboratories frequently encounter samples without any morphological details that make identification difficult. Therefore, determining the source of animal species or breeds from diverse types of samples is vital for the forensic aims of discrimination and recognition of unidentified samples, which represents a key aspect of biodiversity studies (Coyle, 2007).
Another important perspective for species identification is the adulteration of meat which is one of the most important challenges for meat producers, as most of the species used in adulteration are anonymous and difficult to be detected. Hence, more consideration must be paid to the safety and validity of meat products chiefly for public health, legal, and economic worries. Also, religious consideration is an important factor because meat adulteration can be done by whole or partial replacement of high commercial value meat with less expensive options like offal or pork or by adding proteins of different sources (Kamruzzaman et al., 2013). Therefore, the meat manufacturing critically requests techniques that will monitor non-targeted food samples for adulterants to afford evidence of the source and avoid intentional or accidental food samples admixture.
Mitochondrial DNA (mtDNA) polymorphisms have played significant roles in species identification (Moustafa et al., 2017, Awad et al., 2015) together with tracing the origin of specific breeds and the genetic diversity of domestic sheep and other livestock species. The mtDNA has unique properties including maternal inheritance, higher substitution rate, and very lower recombination rate (Ganbold et al., 2019). The control region, also known as the displacement-loop (D-loop) region, is the primary noncoding controlling area for mtDNA replication and transcription. The structure and variation of the mtDNA control region make it conceivable to designate the genetic polymorphisms and maternal source, chiefly since mtDNA shows slight maternal inheritance without reconstruction and with a reasonably fast evolution rate (Hiendleder et al., 1998a). Genetic diversity assessment within and among breeds offers visions into the population construction, relations essential for founding priorities, conservation approaches, genetic enhancement, and ecological use (Ajmone-Marsan et al., 2014).
In Egypt, the total sheep populace is 4.2 million heads, which accounts for 6% of the total meat production. Barki, Ossimi, and Rahmani are the major breeds of sheep, with populations of 470.000, 514.000, and 990.000 respectively (Galal et al., 2005). As indicated by the World Watch List for Domestic Animal Diversity report (FAO, 2003), 1 to 2 breeds disappear each week among the domesticated populations. Here, this circumstance is chiefly disturbing in the developing countries, where quick fluctuations in production systems are prompting cross-breeding or the substitutions of breeds. Hence, this triggers an urgent need to record livestock genetic resources diversity and to plan policies for their sustainable protection (Hanotte & Jianlin, 2006).
Information on the characteristics and genetic diversity of most sheep breeds is limited. Thus, the goal of this study was to apply mtDNA control region sequence (D-Loop) analysis as a tool to evaluate the genetic diversity and phylogeography of four economically important Egyptian sheep breeds. The resultant information can be adopted to formulate national strategies for breeds conservation and sustainable improvement all over the world. Also, this study directed to authenticate sheep species from other taxa to facilitate the determination of the origin of meat from commonly consumed species (cattle, buffalo, goat and pig) and commonly banned species (dog, horse and donkey) for food control and forensic purposes through alignment of the obtained sequences with those existing in the GenBank database.
MATERIALS AND METHODS
This study was approved by the Ethics of Animal Use in Research Committee of Zagazig University and experimental procedures were conducted in accordance with the NIH general guidelines for the Care and Use of Animals in scientific investigations.
Collection of Samples and DNA Extraction
Blood samples were obtained from unrelated sheep of the following four main sheep breeds in Egypt: Rahmani, Ossimi, Barki, and Baladi. A blood sample (5 ml) was drawn aseptically from the jugular veins of each animal into sterilized vacutainer EDTA tubes. Genomic DNA was extracted by the GeneJET whole blood genomic DNA purification mini kit (Fermentas, Thermo Fisher Scientific, USA) as indicated by the producer’s protocol. The extracted DNA was examined for its quality and quantity using 1% agarose gel electrophoresis and spectrophotometric methods, respectively. Intact DNA that demonstrated no smearing was chosen for further examination.
Pcr Amplification and Gel Electrophoresis
A 572-bp fragment of the mtDNA control region from 15,984 bp to 16,556 bp of the sheep mitochondrial genome (access No. AF010406) was amplified by PCR with the following primer sequences: F 5-ACT GCT TGA CCG TAC ATA GTA C-3 and R 5-AGT ATT GAG GAC GGG GTA A-3. PCR amplification reactions were performed in a 20 µl total volume consisting of 10 μl of HotStar Taq® Master mix (Qiagen GmbH, Germany), 1 μl forward primer (10 μM), 1 μl reverse primer (10 μM), 6 μl of RNAse/DNase-free water and 2 μl of DNA template. PCR was done in a T professional thermal cycler (Biometra, Germany). The conditions of cycling included a single initial denaturation at 94°C for 3 min after that 35 cycles of 94°C for 30 sec (denaturation), 56°C for 30 sec (annealing), 72°C for 30 sec (extension) and a last extension step at 72°C for 7 min. The PCR products (10 µl) were separated using 1.5% agarose gel electrophoresis for 20 min at 120 V. A 100-bp DNA ladder (Fermentas, Thermo Fisher Scientific, USA) was used to determine the sizes of the products. The resulting DNA fragments were visualized using UV trans-illumination and analyzed using the BioDoc Analyze gel documentation system (Biometra, Germany).
DNA Purification and Sequencing
DNA fragments were extracted from the gel using a sterile sharp knife and purified via the GeneJET PCR purification kit (Fermentas, Thermo Fisher Scientific, USA), following the manufacturer’s guidelines. Purified products were immediately sequenced by both the forward and reverse primers used for PCR amplification. The sequencing procedure was achieved by the European Custom Sequencing Centre (GATC Biotech AG, Germany).
Sequence Alignment and Phylogenetic Analysis
The sequences were manually amended by Chromas Lite Ver. 2.1.1, (http://technelysium. com.au) and aligned via MEGA 7 software (Kumar et al., 2016). The four analyzed samples sequences were affiliated with reference sequences of diverse haplogroups to outline the haplogroups to which the analyzed samples fitted. The reference sequences were as follows: AF010407, AY829388, DQ852286, and DQ097452 for haplogroup A; DQ852282 and DQ852285 for haplogroup B; DQ097462, DQ097460, DQ852283, and DQ852284 for haplogroup C; DQ852288 and DQ852289 for haplogroup D; and DQ852280 and DQ852281 for haplogroup E. To further establish the evolutionary relationship of Egyptian sheep breeds with other taxa groups, eight mtDNA control region sequences were included for analysis as follows; domestic goat (Capra hircus, KJ420477); desert goat (Capra nubiana, FJ207527); cattle (Bos taurus, AB065126); buffalo (Bubalus bubalis, AF197209); donkey (Equus asinus, KT182635); horse (Equus caballus, AF072975); dog (Canis lupus familiaris, KF574016); and pig (Sus philippensis, DQ779371).
MEGA 7 software was used to evaluate Kimura’s two-parameter genetic distance and create the neighbour-joining (NJ) tree. Bootstrapping with 1000 replicates was used to examine the robustness of the tree.
RESULTS
Pcr Product of the Mtdna D-Loop Region Obtained by Gel Electrophoresis
A 572-bp fragment of the mtDNA D-loop region was amplified in the four sheep breeds via a specific primer sequence (Figure 1). This sequence ranged from 15,984 to 16,557 bp of the complete mitochondrial genome (NCBI GenBank Accession No. GU229278). The obtained sequences were sent to the NCBI GenBank and are presented under accession numbers MF521828-MF521831. Generally, 14 polymorphic sites were noted, of which 10 were parsimony-informative sites, while the lasting 4 were singletons. The average A+T nucleotide composition (56.8%) was higher than that of G+C (43.2%). The nucleotide diversity value was 0.015. Tajima’s neutrality test yielded a value of 1.368.
Pairwise Distance and Phylogenetic Analysis
The genetic distances among the four sheep breeds (Rahmani, Barki, Baladi, and Ossimi) were estimated. The smallest distance was observed between the Baladi and Ossimi breeds (0.000) followed by the distance between the Rahmani and Barki breeds (0.004). Additionally, the analysis revealed a close relationship between sheep and goat breeds. The greatest distance was observed between domestic goat and dog (0.288), followed by the distance between the desert goat and dog (0.270). Small distances were found between the Rahmani, Barki, Baladi, and Ossimi sheep breeds and pig (0.077, 0.081, 0.068, and 0.068, respectively) and buffalo (0.095, 0.099, 0.081, and 0.081, respectively). Additionally, the greatest distance was observed between the sheep breeds and dog (0.258, 0.264, 0.258, and 0.258, respectively). But, the sheep breeds showed less identity with other aligned species (horse and donkey) (Table 1, Figure 2).
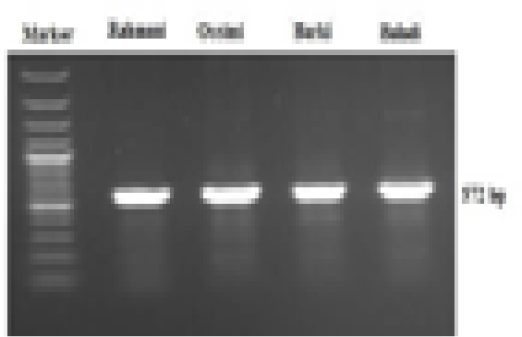
Figure 1: Electrophoretic analysis of PCR product amplified with mtDNA D-loop primer, showing a 572 bp PCR product of four Egyptian sheep breeds mitochondrial genome.
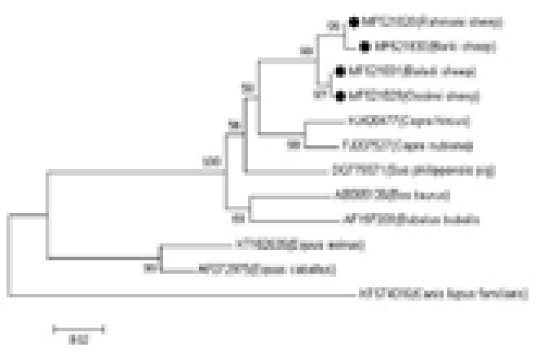
Figure 2: Phylogenetic analysis based on the mtDNA D-loop gene sequences of four Egyptian sheep breeds and other species, showing the evolutionary distance between sheep breeds and other species. The tree was analyzed by neighbor-joining (N-J) analysis.
An NJ phylogenetic tree was plotted by the MEGA 7.0 software (Figure 3). The obtained sequences were aligned with reference sequences of diverse haplogroups to identify the haplogroups to which the analyzed samples fitted. The Ossimi and Baladi breeds clustered with haplogroup A,
Table 1: Genetic distance between four studied Egyptian sheep breeds and its comparison with other species
| Breeds and Species | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
| 1 | MF521828 (Rahmani_sheep) | 1.000 | |||||||||||
| 2 | MF521830 (Barki_sheep) | 0.004 | 1.000 | ||||||||||
| 3 |
MF521831 (Baladi_sheep) |
0.016 | 0.021 | 1.000 | |||||||||
| 4 |
MF521829 (Ossimi_sheep) |
0.016 | 0.021 | 0.000 |
1.000 |
||||||||
| 5 |
KJ420477 (Capra_hircus) |
0.068 | 0.073 | 0.059 | 0.059 | 1.000 | |||||||
| 6 |
FJ207527 (Capra_nubiana) |
0.064 | 0.068 | 0.064 | 0.064 | 0.029 | 1.000 | ||||||
| 7 |
AB065126 (Bos_taurus) |
0.090 | 0.095 | 0.086 | 0.086 | 0.086 | 0.086 | 1.000 | |||||
| 8 |
AF197209 (Bubalus_bubalis) |
0.095 | 0.099 | 0.081 | 0.081 | 0.085 | 0.085 | 0.068 | 1.000 | ||||
| 9 |
KT182635 (Equus_asinus) |
0.177 | 0.182 | 0.188 | 0.188 | 0.204 | 0.198 | 0.187 | 0.182 | 1.000 | |||
| 10 |
AF072975 (Equus_caballus) |
0.162 | 0.167 | 0.172 | 0.172 | 0.182 | 0.177 | 0.172 | 0.182 | 0.042 | 1.000 | ||
| 11 |
KF574016 (Canis_lupus_familiaris) |
0.258 | 0.264 | 0.258 | 0.258 | 0.288 | 0.270 | 0.264 | 0.263 | 0.220 | 0.214 | 1.000 | |
| 12 |
DQ779371 (Sus_philippensis_pig) |
0.077 | 0.081 | 0.068 | 0.068 | 0.067 | 0.067 | 0.081 | 0.094 | 0.177 | 0.157 | 0.264 |
1.000 |
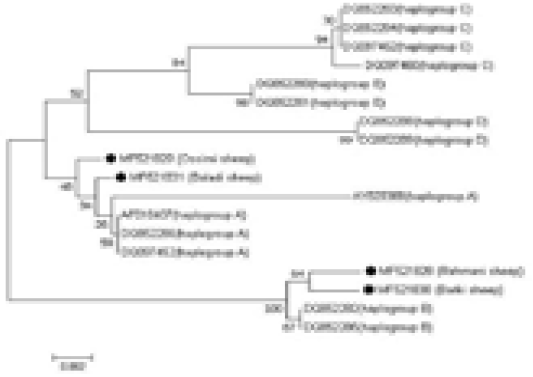
Figure 3: Alignment of the obtained four Egyptian sheep breeds sequences with reference sequences of different haplogroups, showing the haplogroups to which the analyzed samples belonged.
whereas the Rahmani and Barki breeds clustered with haplogroup B. The other haplogroups, i.e., C, D, and E, described in the literature were not found. Additionally, the comparative analysis of the mtDNA control region sequences of the Egyptian sheep breeds with those of the domestic goat, desert goat, cattle, buffalo, donkey, horse, dog, and pig revealed that the Egyptian sheep breeds clustered most closely with goat followed by pig because of their relatedness.
DISCUSSION
Mitochondrial sequencing has been widely utilized by geneticists to elucidate many domestic livestock species origins comprising cattle, swine, goats, and buffalo (Giuffra et al., 2000, Sultana et al., 2003, Tanaka et al., 1996, Zhang et al., 2004). Identification of the species of food animals is viewed as imperative for social, forensic, and public health reasons. Sheep data are starting to coordinate with the pattern detected in other native species (Othman et al., 2015). Two (Hiendleder et al., 2002, Hiendleder et al., 1998b, Wood & Phua, 1996), three (Guo et al., 2005, Pedrosa et al., 2005) and then five (Meadows et al., 2007) ancestries were distinguished in sheep. The chief haplotypes (A and B) are both present in Asia, while B predominates in Europe. Haplotype C has been present in Turkey, Portugal, China, and the Caucasus (Tapio et al., 2006). Haplotype D, which is found in Caucasian animals, and Rumanian Karachai is conceivably identified by the A haplotype. Haplotype E was detected in the Middle East. Five maternal lineages (haplogroups A-E) have been defined in sheep. Haplogroup A and B are of Asian and European sources, respectively. Haplogroups C, D, and E originated in the Near East (Meadows et al., 2007). The mtDNA nucleotide diversity and haplotype are two important tools used to evaluate genetic polymorphisms and distinction (Pereira et al., 2006).
In the current investigation, aside from the A and B haplogroups, no other described haplogroups were noted in the investigated Egyptian sheep population. The Baladi and Ossimi breeds express mainly the A haplotype, while the Rahmani and Barki sheep breeds belong to the B haplogroup, reflecting the close relationship of each pair. Additionally, Gornas et al. (2011) reported the presence of Sudanese sheep in the A and B haplotypes, suggesting a contact zone between Sudanese and Egyptian sheep ancestors. Also, Haplogroup B is dominant Algerian sheep breeds (Ghernouti et al., 2017). This result is consistent with Hiendleder et al. (1998a) who suggested the B haplotype domination in African sheep. Two sheep breeds in the current study were found to belong to the A haplotype. This haplotype is predominantly present in Asian sheep (Hiendleder et al., 1998b). The A haplotype that was detected in populations in Egypt could be owed to partial heritage from an older Asian ancestor presented with the Arabian tribes during their early arrival to Egypt. In contrast to these findings, Chinese and Turkish sheep were detected in 3 haplogroups (Chen et al., 2006, Pedrosa et al., 2005) and sheep from Central Asia were found in five haplogroups (Meadows et al., 2007). Also, in a recent phylogenetic analysis by Ganbold et al. (2019) in Mongolian native sheep, three haplogroups (A, B and C) were identified. In addition, Othman et al. (2015) demonstrated that the most dominant haplogroup recorded in Egyptian breeds was B; however, a few Egyptian animals clustered with the A or C haplogroup. The outcomes of the current study provide basic understanding of the probable source and development of the Egyptian ovine genetic resources.
A Controversy is found concerning the origin of sheep. Using mtDNA PCR-RFLP and sequence analysis methods, previous reports on the origin of domestic sheep from Europe, Africa, New Zealand, and Central Asia found that the urial (O. vignei bochariensis) and argli (O. ammon nigrimontana) species were the main origin species (Hiendleder et al., 1991, Hiendleder et al., 2002, Hiendleder et al., 1998b, Wood & Phua, 1996).
Lately, in several countries, commercially available food products comprising mutton and beef have been found to contain undeclared horse, donkey, and dog meat in addition to other undeclared species. Additionally, nearly whole the meat amount in a few cases was undeclared meat (Brown, 2013, ÖZPINAR et al., 2013). Religious beliefs are another serious factor that should be considered in meat adulteration. The canine meat-eating is illegal in Buddhism and Islam (Rahman et al., 2014). But, pork consumption is prohibited in Judaism and Islam (Nakyinsige et al., 2012). Pork DNA was distinguished in various Halal meats even though it was marked as Halal-certified. Consequently, Muslims and Jews have religious worries about pork consumption and any deviance might lead to reduced consumption together with religious concepts’ violation.
In this study, the close relationship of sheep breeds with pig and the reduced identity with the dog were recorded through comparison of the obtained sequences with other available sequences using NCBI BLAST. In contrast, Rahman et al. (2014) targeted the cytochrome b gene for the recognition of meat adulteration and found that the pair-wise distance was obtained by the maximum complex likelihood method. Tamura et al. (2011) showed a close association between dog and sheep (0.256). A comparable result was noted after phylogenetic tree created by the NJ way (Saitou & Nei, 1987). Thus, the findings of the current investigation are concordant with those of Girish et al. (2005), who employed PCR-RFLP of the mt 12s rRNA gene and established the technique to be valuable for the meat recognition of goat meat, buffalo meat, beef, and sheep. Also, in a recent study of Vaithiyanathan et al. (2018), using DNA based molecular techniques like mt 12 S rRNA PCR assay and mt D loop/cyt b duplex species-specific PCR gave definite results of meat species identification. Consequently, the former studies have revealed the effectiveness of DNA-based techniques in detailed animal species identification (Ali et al., 2015, Rastogi et al., 2007).
Conclusion
Overall, the genetic diversity of Egyptian sheep breeds is low, and evolutionary analysis together with the sequences from GenBank further proposed that Egyptian sheep breeds are originated from two diverse maternal resources. Additionally, distinguishing sheep species from other taxa facilitates identification of the origin of meat from commonly consumed species (cattle, buffalo, goat, and pig) and commonly banned species (dog, horse, and donkey) for food control and forensic purposes.
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
AUTHORS CONTRIBUTION
Yasmina M. Abd- Elhakim designed the study, collected the samples, and drafted the manuscript. Amir H. Abd-Elfatah performed the genetic analysis and revised the manuscript. Walaa M. Elhady designed the study and revised the manuscript. Mayada R. Farag designed the study and revised the manuscript.
REFRENCES






