Advances in Animal and Veterinary Sciences
Research Article
Advances in Animal and Veterinary Sciences 2 (6): 365 – 368Isolation and Identification of Porcine Circovirus 2 from Cases of Respiratory Disease and Postweaning Multisystemic Wasting Syndrome in Pigs
R. Anoopraj1, Jeny K. John1, Menaka Sethi1, R. Somvanshi2, G. Saikumar1*
- National CSF Referral Laboratory and Swine Disease Laboratory, Division of Pathology, Indian Veterinary Research Institute, Izatnagar, Uttar Pradesh, India
- EBH Laboratory, Division of Pathology, Indian Veterinary Research Institute, Izatnagar, Uttar Pradesh, India
*Corresponding author: saikumarivri@gmail.com
ARTICLE CITATION:
Anoopraj R, John JK, Sethi M, Somvanshi R, Saikumar G (2014). Isolation and identification of porcine circovirus 2 from cases of respiratory disease and postweaning multisystemic wasting syndrome in pigs. Adv. Anim. Vet. Sci. 2 (6): 365 – 368.
Received: 2014–06–11, Revised: 2014–07–10, Accepted: 2014–07–11
The electronic version of this article is the complete one and can be found online at
(
http://dx.doi.org/10.14737/journal.aavs/2014/2.6.365.368
)
which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited
ABSTRACT
Porcine circovirus disease (PCVD), an emerging viral disease of economic importance, represents several disease manifestations principally caused by porcine circovirus 2 (PCV2). Prevalence of distinct PCV2 genotypes in pig population necessitates to study their replication potential in cell line in order to establish and compare infectivity and pathogenecity. Virus isolation forms an important aid in PCVD diagnosis, especially when multifactorial origin of disease condition is suspected. In the present study, isolation of PCV2 was accomplished by serially passaging the inoculum prepared from suspected tissue samples up to fourth passage in PK–15 cell line. Further, the confirmation of replication in cell line was done by immunoperoxidase monolayer assay and polymerase chain reaction. Information regarding virus isolation and identification of PCV2 isolates circulating in Indian pigs is scarce. In the present work, PCV2 was successfully isolated in PK–15 cell line and demonstrated their replication in cytoplasm and in nucleus by immunoperoxidase monolayer assay. Thus, virus isolation together with PCR assay helped to establish the PCV2 etiology in pneumonia and wasting of pigs encountered in the present study.
INTRODUCTION
Porcine circovirus disease (PCVD) is an important emerging viral disease in India with worldwide occurrence inflicting heavy economic losses to swine industry (Ellis et al., 1999; Sharma and Saikumar, 2010; Meng, 2012; Rajkhowa and Saikumar, 2012; Tico et al., 2013). Porcine circovirus type 2 (PCV2), principal causative agent of PCVDs, is associated with several clinical conditions like PCV2 systemic disease (PCV2–SD) which includes post weaning multisystemic wasting syndrome (PMWS) , PCV2 lung disease (PCV2–LD), PCV2 enteric disease (PCV2–ED), PCV2 reproductive disease (PCV2–RD) and porcine dermatitis and nephropathy syndrome (PDNS) (Segales, 2012). Usually, PCVDs affect pigs of 5–18 weeks age with morbidity rate of 4–60% and mortality rate of 4–20% (Segales and Domingo, 2002; Zhu et al., 2007).
PCV2 belongs to the family circoviridae, genus circovirus, is a small, icosahedral, non–enveloped, single stranded DNA virus containing a circular genome of 17 nm diameter and with 1767–1768 nucleotides (Hamel et al., 1998; Dupont et al., 2008). The PCV2 genome has three major open reading frames (ORFs). PCV2 ORF1 (945 bp) gene encodes replication proteins Rep and Rep’ involved in virus replication (Mankertz et al., 1997). ORF2 (702 or 705 bp), encodes the major capsid protein (Cap) which is also the main antigenic determinant of the virus (Nawagitgul et al., 2000). Both ORF 1 and ORF 2 together comprise 93 % of the PCV2 genome (Opriessnig et al., 2006). ORF3 encodes a protein that is not essential for PCV2 replication, but involved in PCV2–induced apoptosis (Liu et al., 2005).
PCV2 can be divided into three distinct genotypes, namely PCV2a, PCV2b, and PCV2c, by phylogenetic analysis based on ORF2 gene and complete genome. Infectivity and pathogenecity of these genotypes vary and PCV2b is considered as the most virulent genotype (Olvera et al., 2007). Recently, emergence of new variants of PCV2 has also been reported that evolved through events of mutation and/or recombination (Ramos et al., 2013). Therefore, to establish infectivity and pathogenecity, it is essential to demonstrate their replication potential in vitro.
Diagnosis of PCVDs is accomplished chiefly by clinical signs, characteristic gross and histopathological findings and demonstration of viral nucleic acid or antigen in associated lesions. Though, virus isolation is not a gold standard method for PCVDs diagnosis, it adds confidence in confirmation of diagnosis especially when multiple pathogens are involved. PCV–free PK–15 cell line is considered to be the best cell line for replication of PCV2 and is used widely for virus isolation (Tischer et al., 1987; Zhu et al., 2007). Absence of cytopathic effects after infection necessitates demonstration of viral nucleic acid or protein in the infected cell line to monitor viral replication (Allan and Ellis, 2000). Herein, we report the isolation and identification of PCV2 in PK–15 cell line from cases of PCV2–SD with prominent respiratory involvement and PCV2–SD associated with PMWS.
MATERIALS AND METHODS
Collection of Samples
Tissue samples of lung, lymph node, spleen, tonsil and liver were collected aseptically from animals died of pneumonia and PMWS associated PCV2–SD case with respiratory signs and wasting. Animal died of pneumonia was a suckling piglet of 3 days age, which showed non–collapsed lungs with cranio–lateral and ventral consolidation of right intermediate lobe and severely congested carcass lymph nodes at necropsy, while the animal with PMWS was a grower that showed progressive wasting in spite of normal feed intake. At necropsy this animal revealed confluent consolidation of lung lobes, randomly distributed small circumscribed abscesses in right diaphragmatic lobe and severely congested carcass lymph nodes. Samples were transported on ice to the laboratory and stored at –800C till further processing.
PCV2 Detection
DNA was isolated from collected samples of lung, lymph node and spleen of each case by using QIAmp DNA Mini and Blood Mini kit (Qiagen, USA) following manufacturer’s instructions. Initial screening was performed by PCR targeting a fragment of 481 bp size corresponding to ORF2 region of PCV2 (Ellis et al., 1999).
Virus Isolation
Twenty four hours grown, 50–80% confluent PCV–free PK–15 cell cultures were used for inoculation with infected samples. The procedure for PCV2 isolation was adopted as described by Wellenberg et al., (2000) with minor modifications and is outlined as follows. Inoculum for infection of cell line was prepared and consisted of pooled sample of lung, lymph node and liver of necropsy no. 89A–13 and 192A–13. Five hours after inoculation of PK–15 cell line with the prepared inoculum, semi–confluent monolayer was treated with 300 mM D–glucosamine–HCl and incubated for 30 min followed by washing twice with DMEM to remove the D–glucosamine–HCl and then incubation in DMEM containing 10% FCS and 0.5% antibiotic mix for further 48 hrs at 370C in a 5% CO2 incubator. Negative controls were processed in the same way. After incubation period, cell cultures were subjected to freeze–thaw cycles three times (frozen at –700C for one hour followed by thawing). The resultant cell lysate was centrifuged at 2,500 g and the supernatant was again passaged. After each passage, the freshly seeded flasks were incubated for 24 hrs, treated with D–glucosamine–HCl, and further incubated for 48 hrs as described above. After four passages, the monolayer of PK–15 was screened for PCV2 antigen by immunoperoxidase test and nucleic acid by PCR.
Detection of PCV 2 Replication by Indirect Immunoperoxidase Monolayer assay (IPMA)
For detection of PCV 2 replication by immune labelling with HRPO detection system, cover slip monolayer preparations of PK–15 cell cultures infected with fourth passage suspension prepared in 6 well plates were used. After the incubation for 48 hrs, the cover slips were harvested, fixed with acetone/methanol (1:1) solution for 10 min at RT. The cover slips were drained thoroughly and dried under a bench lamp for 4 hrs at 25–30oC. Fixed preparations were stored at – 20oC in a sealed bag before staining.
The immune labelling procedure is briefly outlined as follows. The coverslips were brought to RT, mounted onto glass slides and rinsed once in PBS for 5 min. Subsequently endogenous peroxidase quenching was done with 3% hydrogen peroxide in methanol followed by blocking with 5% normal goat serum in PBS. Porcine Circovirus 2 antiviral anti–serum (polyclonal sera, used 1:250 in PBS, VMRD, USA) was used as primary antibody, while goat anti–pig IgG conjugated with HRPO enzyme (1:500 dilution in PBS; Bethyl, USA) was used as secondary antibody. After incubation with primary and secondary antibody, AEC (3–Amino–9–ethylcarbazole) chromogen solution was applied to the monolayer preparation for 10 min with continuous monitoring under a microscope. After satisfactory development of colour reaction, the reaction was stopped by rinsing the slides gently with distilled water and counterstained with Mayer’s haematoxylin for 2 min.
Detection of PCV2 Replication by PCR
Control as well as virus inoculated flasks containing fourth passage cells were freeze thawed three times. 2 ml of the cell lysate from each flask was used for DNA extraction. The harvested cell lysates were centrifuged at 13,000 rpm for 5 min at RT and the DNA was extracted from the resultant supernatant as well as cell sediment by QIAmp DNA Mini and Blood Mini kit. Later, PCR was carried out to confirm the replication of PCV2 with PCV2 2F (5’–CGG ATA TTG TAG TCC TGG TCG–3’) and PCV2 2R (5’–ACT GTC AAG GCT ACC ACA GTC A–3’) primer set, amplifying a 765 bp fragment. PCR reactions were performed in 25 µL reaction volume containing 10X PCR buffer, 50 mM MgCl2, 0.2 mM of each dNTP, 10 µM of each primer and 1.5 U Taq DNA polymerase (Maxima Hot Start Taq DNA polymerase, Thermo Scientific). PCR reaction conditions were 950C for 4 min, followed by 40 cycles of 950C for 30s, 620C for 40s, 720C for 1 min, and final extension step at 720C for 7 min. DNA extracted from control cells served as negative control template, while a known positive (192A–13, lymph node) sample served as positive control. After electrophoresis, the amplified products were viewed under UV imaging system.
RESULTS
PCV2 Detection
During initial screening, twelve cases were found positive for PCV2 by PCR. Samples from two positive cases (Necropsy no. 89A–13 and 192A–13) were selected for virus isolation based on relative freshness of the sample.
Virus Isolation
After inoculation with the samples, the cells exhibited a slow growth compared to the negative uninfected control cells. No cytopathic effect was observed. Morphologically, there were no differences in the infected and uninfected cells, except for the presence of mild to moderate cytoplasmic vacuolation in the infected cells after fourth passage (Figure 1). The virus could be successfully isolated
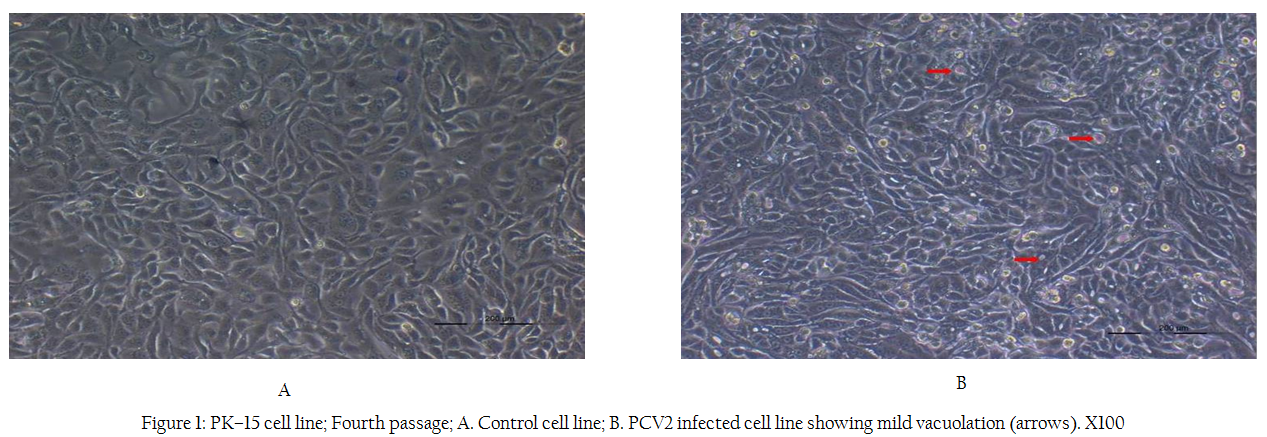
Figure 1: PK–15 cell line; Fourth passage; A. Control cell line; B. PCV2 infected cell line showing mild vacuolation (arrows). X100
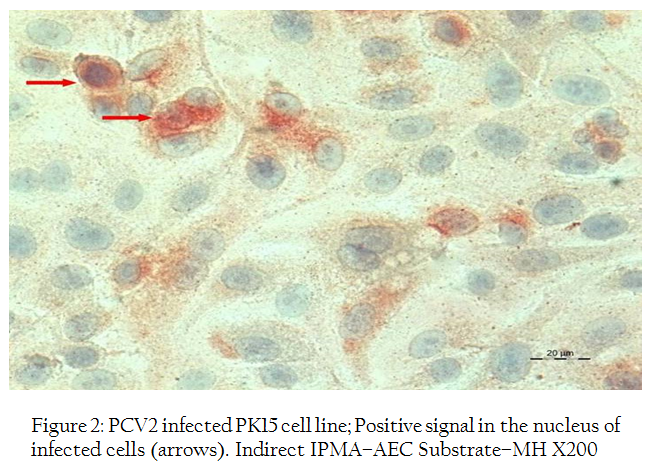
Figure 2: PCV2 infected PK15 cell line; Positive signal in the nucleus of infected cells (arrows). Indirect IPMA–AEC Substrate–MH X200
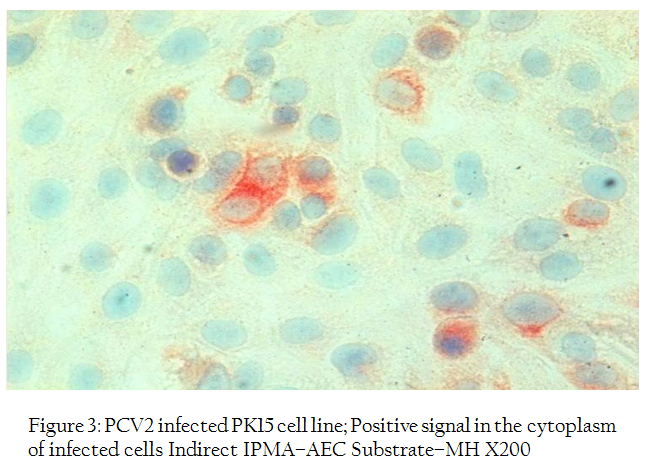
Figure 3: PCV2 infected PK15 cell line; Positive signal in the cytoplasm of infected cells Indirect IPMA–AEC Substrate–MH X200
Indirect Immunoperoxidase Monolayer Assay (IPMA) and PCR
Indirect immunoperoxidase test of PCV2 infected PK–15 monolayer using porcine circovirus 2 antiviral anti–serum revealed both intracytoplasmic and intranuclear positive reaction (Figures 2, 3). In a few cells, positive reactions were present in the entire nucleus of the cell. The replication of virus was further confirmed by PCR reaction with the specific primers which showed positive band of expected size of 765 bp for both cell culture supernatant and cell sediment DNA (Figure 4).
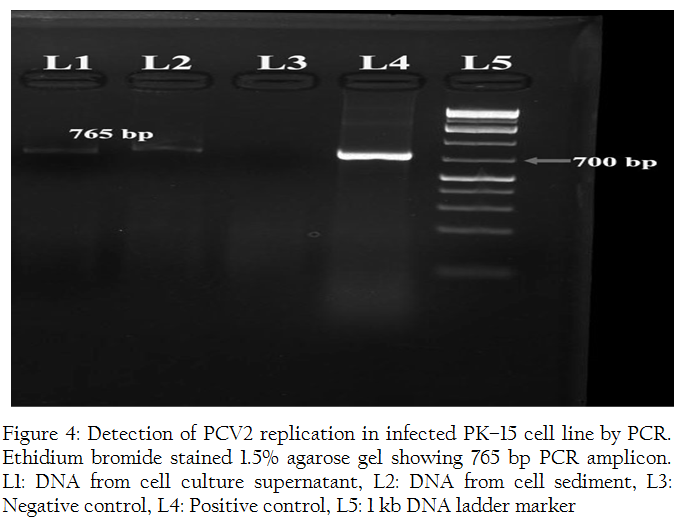
Figure 4: Detection of PCV2 replication in infected PK–15 cell line by PCR. Ethidium bromide stained 1.5% agarose gel showing 765 bp PCR amplicon. L1: DNA from cell culture supernatant, L2: DNA from cell sediment, L3: Negative control, L4: Positive control, L5: 1 kb DNA ladder marker
DISCUSSION
PCV2 is ubiquitous in nature. The virus can be recovered from both healthy as well as diseased animals (Jantafong et al., 2011). Therefore, the diagnosis PCVDs requires additional information from clinical signs and associated lesions and mere PCR positivity cannot establish the disease condition. However, PCR is a rapid and reliable technique to detect PCV2 infections (clinical and subclinical) in pigs (Alarcon et al., 2011). Thus, in our study, we chose PCR as detection method to identify PCV2 infections.
Virus isolation is the most reliable method to establish an infection of viral etiology and is considered as the cornerstone in laboratory diagnosis of viral infections (Storch, 2000). In the present study, PCV2 was successfully isolated in PK–15 cell line after infecting the cells with a pooled inoculum of lung, lymph node and spleen from two PCR positive cases and thus confirmed a PCV2 etiology in the disease condition. Treatment of PK–15 cell line with 300 mM D–glucosamine–HCl is recommended after infection with PCV2 to enhance viral replication. This is because, D–glucosamine–HCl enables PCV2 to enter nucleus of PK–15 cells, wherein the viruses utilise the cellular enzymes expressed in S–phase of cell cycle for their replication (Tischer et al., 1987). However, prolonged treatment with D–glucosamine–HCl can lead to cellular damage due to its toxic effects on cell cultures (Allan and Ellis, 2000). After inoculation, the infected cell line showed a slow rate of growth compared to negative control. Virus replication in cell line did not produce any cytopathic effect. This is consistent with previous studies that described similar findings (Wellenberg et al., 2000; Rajkhowa, 2008). A positive indirect immunoperoxidase test indicated the replication of PCV2 in cell culture. The staining pattern in the infected cells was similar to the pattern described by Allan et al. (1998). Identification of intracytoplasmic and intranuclear staining revealed the presence of viral antigen in both cell compartments. In addition, a positive PCV2 specific amplification of DNA isolated from fourth passage cells and supernatant by PCR further underlined PCV2 replication in cell line. Though, virus isolation is not considered as the gold standard method for PCVD diagnosis due to absence of cytopathic effect, it adds confidence to the diagnosis of PCVD in combination with IPMA and PCR (Wellenberg et al., 2000; Kim and Chae, 2004). This is particularly important in establishing the role of PCV2 as a pathogen in condition like PCV2–SD, since its clinical manifestation is widely considered as a multifactorial outcome. Moreover, in a study conducted to compare efficiency of various diagnostic methods to detect PCV2, virus isolation was found to be more sensitive than immunohistochemistry and in situ hybridisation for demonstration of the virus (Kim and Chae, 2004).
Therefore, the successful isolation of PCV2 in PK–15 cell line in the present study helped in confirming the disease etiology. Likewise, isolation studies pertaining to different genotypes of PCV2 help in identifying the differences in their pathogenecity. This will be much beneficial in identifying the replication potential and infectivity of newly emerging recombinant strains of PCV2.
ACKNOWLEDGEMENT
The authors thank Department of Biotechnology, Govt. of India, for their financial support and acknowledge the Director, Indian Veterinary Research Institute for providing facilities to conduct this research work.
CONFLICT OF INTEREST
No conflict of interest to declare.
REFERENCES
Alarcon P, Velasova M, Werling D, Stark KD, Chang YM, Nevel A, Pfeiffer DU, Wieland B (2011). Assessment and quantification of post–weaning multi–systemic wasting syndrome severity at farm level. Prev. Vet. Med. 98: 19 – 28.
http://dx.doi.org/10.1016/j.prevetmed.2010.09.022
PMid:21036410
Allan GM, Ellis JA (2000). Porcine circoviruses: a review. J. Vet. Diagn. Invest. 12: 3 – 14.
http://dx.doi.org/10.1177/104063870001200102
PMid:10690769
Allan GM, McNeilly F, Kennedy S, Daft B, Clarke EG, Ellis JA, Haines DM, Meehan BM, Adair BM (1998). Isolation of porcine circovirus–like viruses from pigs with a wasting disease in the USA and Europe. J. Vet. Diagn. Invest. 10: 3 – 10.
http://dx.doi.org/10.1177/104063879801000102
PMid:9526853
Dupont K, Nielsen EO, Baekbo P, Larsen LE (2008). Genomic analysis of PCV2 isolates from Danish archives and a current PMWS case–control study supports a shift in genotypes with time. Vet. Microbiol. 128: 56 – 64.
http://dx.doi.org/10.1016/j.vetmic.2007.09.016
PMid:17996404
Ellis JA, Krakowka S, Lairmore M, Haines DM, Bratanich A, Clark EG, Allan GM, Konoby C, Meehan BM, Kennedy S, McNeilly F (1999). Reproduction of lesions of postweaning multisystemic wasting syndrome in gnotobiotic piglets. J. Vet. Diagn. Invest. 11: 3 – 14.
http://dx.doi.org/10.1177/104063879901100101
PMid:9925205
Hamel AL, Lin LL, Nayar GP (1998). Nucleotide sequence of porcine circovirus associated with postweaning multisystemic wasting syndrome in pigs. J. Virol. 72: 5262 – 5267.
PMid:9573301 PMCid:PMC110114
Jantafong T, Boonsoongnern A, Poolperm P, Urairong K, Lekcharoensuk C, Lekcharoensuk P (2011). Genetic characterization of porcine circovirus type 2 in piglets from PMWS–affected and –negative farms in Thailand. Virol. J. 8: 88.
http://dx.doi.org/10.1186/1743-422X-8-88
PMid:21356069 PMCid:PMC3058092
Kim J, Chae C (2004). A comparison of virus isolation, polymerase chain reaction, immunohistochemistry, and in situ hybridization for the detection of porcine circovirus 2 and porcine parvovirus in experimentally and naturally co–infected pigs. J. Vet. Diagn. Invest. 16: 45 – 50.
http://dx.doi.org/10.1177/104063870401600108
PMid:14974846
Liu J, Chen I, Kwang J (2005). Characterization of a previously unidentified viral protein in porcine circovirus type 2–infected cells and its role in virus induced apoptosis. J. Virol. 79: 8262 – 8274.
http://dx.doi.org/10.1128/JVI.79.13.8262-8274.2005
PMid:15956572 PMCid:PMC1143760
Mankertz A, Persson F, Mankertz J, Blaess G, Buhk HJ (1997). Mapping and characterization of the origin of DNA replication of porcine circovirus. J. Virol. 71: 2562 – 2566.
PMid:9032401 PMCid:PMC191374
Meng XJ (2012). Spread like a wild fire–The omnipresence of porcine circovirus type 2 (PCV2) and its ever expanding association with diseases in pigs. Virus Res. 164: 1 – 3.
http://dx.doi.org/10.1016/j.virusres.2011.12.005
PMid:22192532
Nawagitgul P, Morozov I, Bolin SR, Harms PA, Sorden SD, Paul PS (2000). Open reading frame 2 of porcine circovirus type 2 encodes a major capsid protein. J. Gen. Virol. 81: 2281 – 2287.
PMid:10950986
Olvera A, Cortey M, Segales J (2007). Molecular evolution of porcine circovirus type 2 genomes: phylogeny and clonality. Virology. 357: 175 – 185.
http://dx.doi.org/10.1016/j.virol.2006.07.047
PMid:16963096
Opriessnig T, McKeown NE, Zhou EM, Meng XJ, Halbur PG (2006). Genetic and experimental comparison of porcine circovirus type 2 (PCV2) isolates from cases with and without PCV2–associated lesions provides evidence for differences in virulence. J. Gen. Virol. 87: 2923 – 2932.
http://dx.doi.org/10.1099/vir.0.82099-0
PMid:16963751
Rajkhowa TK (2008). Studies on pathology and diagnosis of PCV2 associated diseases. Ph.D. Thesis (Vety. Pathology), Deemed University, IVRI, Izatnagar, UP.
Rajkhowa TK, Saikumar G (2012). Occurrence of post weaning multisystemic wasting syndrome in crossbred pigs of Uttar Pradesh and Uttarakhand, India. Indian J. Anim. Sci. 82: 1122 – 1127.
Ramos N, Mirazo S, Castro G, Arbiza J (2013). Molecular analysis of porcine circovirus type 2 strains from Uruguay: Evidence for natural occurring recombination. Infect. Genet. Evol. 19: 23 – 31.
http://dx.doi.org/10.1016/j.meegid.2013.06.017
PMid:23806516
Segales J (2012). Porcine circovirus type 2 (PCV2) infections: Clinical signs, pathology and laboratory diagnosis. Virus Res. 164: 10 – 19.
http://dx.doi.org/10.1016/j.virusres.2011.10.007
PMid:22056845
Segales J, Domingo M (2002). Postweaning multisystemic wasting syndrome (PMWS) in pigs. A review. Vet. Q. 24: 109–124.
http://dx.doi.org/10.1080/01652176.2002.9695132
PMid:12400999
Sharma R, Saikumar G (2010). Porcine parvovirus and porcine circovirus 2–associated reproductive failure and neonatal mortality in crossbred Indian pigs.Trop. Anim. Health Prod. 42: 515 – 522.
http://dx.doi.org/10.1007/s11250-009-9454-0
PMid:19763866
Storch GA (2000). Diagnostic virology. Clin. Infect. Dis. 31: 739 – 751.
http://dx.doi.org/10.1086/314015
PMid:11017824
Tico G, Segales J, Martinez J (2013). The blurred border between porcine circovirus type 2–systemic disease and porcine respiratory disease complex. Vet. Micrbiol. 163: 242 – 247.
http://dx.doi.org/10.1016/j.vetmic.2013.01.001
PMid:23398668
Tischer I, Peters D, Rasch R, Pociuli S (1987). Replication of porcine circovirus: induction by glucosamine and cell cycle dependence. Arch. Virol. 96: 39 – 57.
http://dx.doi.org/10.1007/BF01310989
PMid:3619654
Wellenberg GJ, Pesch S, Berndsen FW, Steverink PJGM, Hunneman W, Van der Vorst TJK, Peperkamp NHMT, Ohlinger VF, Schippers R, Van Oirschot JT, de Jong MF (2000). Isolation and characterization of porcine circovirus type 2 from pigs showing signs of post–weaning multisystemic wasting syndrome in the Netherlands. Vet. Q. 22: 167 – 172.
http://dx.doi.org/10.1080/01652176.2000.9695049
PMid:10952449
Zhu Y, Lau A, Lau J, Jia Q, Karuppannan AK, Kwang J (2007). Enhanced replication of porcine circovirus type 2 (PCV2) in a homogeneous subpopulation of PK15 cell line. Virology 369: 423 – 430.
http://dx.doi.org/10.1016/j.virol.2007.08.014
PMid:17889922





