Advances in Animal and Veterinary Sciences
Research Article
Molecular Identification of Theileria equi and Babesia caballi from Ixodid Ticks Infesting Equids in Erbil Province, Northern of Iraq
Khalid Jabar Aziz1*, Lokman Taib Omer AL-Barwary2
1Department of Animal Resources, College of Agricultural Engineering Sciences, Salahaddin University-Erbil; 2Department of Microbiology and Pathology, College of Veterinary Medicine, University of Duhok.
Abstract | The present study was conducted to detect of Theileria equi and Babesia caballi in Ixodid ticks collected from equids in Erbil province, Northern Iraq. A total of (98) adult ticks, belong to the five species from three ixodid genera (two species belong to the genus Hyalomma (Hyalomma marginatum marginatum and H. anatolicum exacavatum), two species belong to the genus Boophilus (Boophilus microplus and B. annulatus) and one species belong to the genus Rhipicephalus (Rhipicephalus turanicus) were collected from 349 equids. Fifty DNA ticks (10 for each tick species) were chosen for molecular analyses to detect of T. equi and B. caballi using conventional and multiplex polymerase chain reaction technique (c-PCR, m-PCR) targeting 18S rRNA gene. Out of analyzed ticks, Boophilus annulatus was the most prevalent tick species (39.8%), whereas the lowest was Hyalomma exacavatum (10.2%). The multiplex PCR finding revealed that five ticks species including (Hyalomma marginatum marginatum, Hyalomma anatolicum exacavatum, Boophilus microplus, Boophilus annulatus and Rhipocephalus turanicus) were infected with T. equi, while only one species Rhipocephalus turanicus and Boophilus annulatus was found infected with Babesia caballi and both protozoa respectively.
Keywords | Ixodid, Theileria equi and Babesia caballi, PCR, Iraq
Received | July 17, 2020; Accepted | September 04, 2020; Published | November 15, 2020
*Correspondence | Khalid Jabar Aziz, Department of Animal Resources, College of Agricultural Engineering Sciences, Salahaddin University-Erbil; Email: khalid.aziz1@su.edu.krd
Citation | Aziz, KJ, AL-Barwary LTO (2020). Molecular identification of Theileria equi and Babesia caballi from ixodid ticks infesting equids in Erbil province, northern of Iraq. Adv. Anim. Vet. Sci. 8(12): 1286-1293.
DOI | http://dx.doi.org/10.17582/journal.aavs/2020/8.12.1286.1293
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2020 Aziz and AL-Barwary. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
Ixodid ticks are important ectoparasite due to their capacity to harbor and transmit a variety of zoonotic and veterinary important pathogens including (Protozoa, bacteria, virus and rickettsia) to domestic and wild animals in most region of the world (Ozdarendeli et al., 2008; Aziz and AL-Barwary, 2019a). Different diseases can be associated with this parasite and namely tick-borne disease (TBDs) (Otranto et al., 2014). In past twenty years, countless TBDs have been recognized in various countries (Kernif et al., 2016). Eight main species of ixodid ticks including (Rhipicephalus turanicus, R. sanguines, Hyalomma anatolicum anatolicum, H. marginatum marginatum, H. anatolicum excavatum, Boophilus annulatus, H, turanicum and H detritum detritum) were previously recorded from cattle, sheep and goats in Erbil region, Northern Iraq (Ameen, 2012). Babesia caballi and Theileria equi are the most important tick-borne protozoal pathogens causing equine piroplasmosis (EP) infecting the equidae family (horses, mules, donkeys, ponies and zebras) in which it cause significant hygienic and economic losses in horses (Manna et al., 2018). Babesia caballi and Theileria equi have been classified taxonomically in the phylum Apicomplexa (protozoa) and in two different families, Babesiidae and Theileriidae, respectively (Taylor et al., 2007). Despite the fact that Theileria equi are more prevalent and more pathogenic than Babesia caballi in endemic areas, these two parasite always share the same tick vectors to infest equids (Katz, et al., 2000; Sigg et al., 2010). The protozoa are mainly transmitted by ticks of the Ixodidae family (Zangana and Naqid, 2011). Transmission through the tick vectors occurs by two main routs first; mechanical transmission when the mouthparts are contaminated, second; biological transmission when the protozoa develop or proliferate itself in the tick vector (Scoles and Ueti, 2015). The regional distribution of equine piroplasmosis greatly affected by the presence of ticks and international horse trade between endemic and sensitive area (Friedhoff and Tenter,1990; Bruning, 1996). It has been reported that in the areas where tick vectors are commonness, the outbreak of the disease usually associated with entry of the persistently carrier horses (Sumbria et al., 2015). Ticks belong to the family Ixodidae, called hard or Ixodid ticks (Acari: Arachnida), which obligate blood sucking ectoparasites of various mammals, vertebrates, birds, amphibians and reptiles (Schmidt and Roberts, 1989). There is an obligatory relationship between piroplasms and their Ixodid tick vectors (Gou et al., 2013). Out of the 867 tick species currently known, there are 33 Ixodid ticks species belong to six genera that have been reported as competent tick vectors for equine piroplasmosis (Jongejan and Uilenberg, 2004). The Ixodid tick species include; Amblyomma spp., Hyalomma spp., Rhipicephalus spp., Dermacentor spp., Ixodes spp., and Haemaphysalis spp. these ticks have been reported as a risk factors to transmit B. caballi and T. equi to equids (Jongejan and Uilenberg, 2004). Twenty-seven tick species belong to these six genera, can be transmitted T. equi includes Ambyomma spp., Dermacentor spp., Haemaphysalis spp., Hyalomma spp., Ixodes spp., and Rhipicephalus spp. (Scoles and Ueti, 2015). Twenty-four tick species belong to the four genera can transmit B. caballi including Dermacentor spp., Haemaphysalis spp., Hyalomma spp., and Rhipicephalus spp. (Scoles and Ueti, 2015). Ixodid ticks are the final hosts as well as transports for T. equi and B. caballi, due to the former parasites which undergo sexual-stage development in the tick vectors to complete their life cycle (Zapf and Schein, 1994). The most important program to prevent the spreading of the disease is controlling or removing the tick which is considers the main risk factors for EP (Kouam et al., 2010; Guidi et al., 2015).
Molecular tools have been considered as a perfect technique for detection of many species of Babesia and Theileria among different animals in the world. Seemingly, PCR techniques have higher sensitivity and specificity compared with other techniques including serological assays and Giemsa stained blood smears (Buling et al., 2007; Jefferies et al., 2007) Studies have been done to determine the prevalence of equine piroplasmosis from different parts of Iraq using serological techniques and microscopic examination (Alsaad et al., 2012). However, there is a very few molecular epidemiological study on piroplasm parasites from infected animals in Erbil province (Aziz et al., 2019).
The status of EP in ixodid tick among equines has not been evaluated in Erbil province, North of Iraq yet. Therefore, the current study aimed to use conventional and multiplex PCR techniques for the detection of T. equi and B. caballi parasite on tick’s DNA samples collected from equines distributed in different district of Erbil, Northern Iraq.
MATERIALS AND METHODS
Study area and sampling
This study was performed from April 2016 to March 2017 in Erbil province, North of Iraq. A total of 349 equids (Horse, Mule, Donkey and Pony) were inspected for tick infestations. Different part of host body such as head, ears, neck, pectoral, per femoral region, tests, legs, back and buttock, inguinal and under tail areas, were inspected carefully for detection of any tick. A total of 98 ticks of both sexes (male, female) were collected in plastic vials containing 70% ethanol and stored at 4ºC until morphological identification of ticks and molecular identification of EP from tick species by m-PCR.
Identification of ixodid ticks
The species and sex of the collected ticks was identified by studying the morphological characteristics microscopically according to the identification keys (Estrada-Peña et al., 2004). After appropriate identification, all identified ticks were kept individually in 70% ethanol before being treated for further analysis.
DNA extraction from ixodid ticks and PCR amplification
DNA was purified from 50 Ixodid adult ticks (10 ticks) from each species after morphological identification which included: Hyalomma marginatum, Hyalomma anatolicum exacavatum Rhipicephalus turanicus, Rh. (Boophilus) microplus, Rh. (Boophilus) annulatus, using PrimePrepTM Genomic DNA Extraction kit (from Tissue) (GeNet Bio, Korea) according to the manufacturer’s instructions.
In this study, two types of PCR reaction were applied: firstly, to identify the positive equine for all possible Babesia spp., and Theileria spp., the universal ‘catch-all’ primers (Bec-UF1and Bec-UR) by c-PCR assay were used to amplify DNA fragments Secondly, to differentiate between B. caballi and T. equi in all positive and negative samples in the first reaction, by using a single forward primer (Bec-UF2) and two reverse primers, one for identifying B. caballi and another for T. equi, which were Cab-R and Equi-R respectively by m-PCR assay (Table 1).
A m-PCR was performed according to the method of Alhassan et al. (2005) and Qablan et al. (2013) with some modifications. Briefly, 25 µӀ of a mixture containing 2 µӀ of template DNA, 1 µӀ (10 pmol) of each of the reverse primers (EquiR) for T. equi and (CabR) for B. caballi 2 µӀ of a universal forward primer (UF2) , 6.5 dH2O and 12.5 µӀ of 2X PCR master-mix ready to use (GeNet Bio. Laboratory. Korea) with a final concentration of 0.5 mM of each dNTP in 10 mM Tris-HCl, pH 9.0, 4 mM MgCl2,enzyme stabilizer, loading dye and 1 U Taq DNA polymerase. The mixtures were subdue to the following cycling conditions using a BioRad thermocycler: 96oC for 10 min, with 35 cycle of a denaturing step at 96oC for 1 min, an annealing step at 54oC for 1 min, and an extension step at 72oC for 1 min, followed by a final extension at 72oC for 10 min.
After amplification success, a 5 µӀ of the amplificons were runed with gel electrophoresis in a 1.5% agarose for 45 min, then visualised by UV illuminator (Proxima 2500 Isogene Life science, Netherland). DNA positive controls for T. equi and B. caballi were confirmed by nucleotide sequencing and submission to gene bank with accession number MH047214 and MH047215 respectively. Further, DNA extracted from piroplasms-free ticks was used as negative control for PCR amplification.
DNA sequencing and phylogenic analysis
In this study a total of 9 PCR amplicons (8 for T. equi and 1 for B. caballi) that were positive and have strong bands by using m-PCR send to the commercial company for purificated and sequencing (Macrogen Inc. South Korea).
The sequences similarity analyses with previous sequences published in GenBank were performed using the BLAST programme (http://www.ncbi.nlm.nih.gov/BLAST). BLAST analysis approved that all sequences were for T. equi and B. caballi 18S rRNA gene with 97-100 identities to previous published T. equi and B. caballi.
The four DNA samples that represent in Erbil province were applied in the phylogenic analysis. Multiple sequence alignment was done employing MEGA7 version (http://www.megasoftware.net, July 2016). Sequences were aligned together with 18S rRNA gene of T.equi and B. caballi sequences, derived from GenBank, using the Muscle software (Edgar, 2004).
Three different algorithms: Maximum Likelihood, Neighbor-Joining were used to generate a phylogenetic tree and yielded topology-similar results (Tamura-Nei model) with 1000 Bootstrap replicates were performed to estimate the node reliability.
Statistical analysis
Data were analyzed using SPSS from windows (Version 7). Chi-square and Z test were done to find significant differences between tick’s species and sex. A value of p < 0.05 was considered as statistically significant.
RESULTS AND DISCUSSION
Prevalence of Ixodid tick infesting equine in Erbil province
The results show that five species from three ixodid genera (two species belong to the genus Hyalomma (Hyalomma marginatum marginatum and H. anatolicum exacavatum), two species belong to the genus Boophilus (Boophilus microplus and B. annulatus) and one species belong to the genus Rhipocephalus (Rhipocephalus turanicus) were observed on equids during the study. It was found that the tick species B. annulatus (39.8%) was the most frequently observed in equines, while Hyalomma exacavatum (10.2%) showed the minimum occurrence of tick species were observed. Whether as infestation rate of Rhipocephalus turanicus, Boophilus microplus and Hyalomma marginatum were 21.4%, 16.3% and 12.2% respectively Table 2.
Molecular findings
For the first reaction finding of PCR amplified product using universal ‘catch-all’ primers (Bec-UF1and Bec-UR) visualised that the DNA bands size was 876 bp and 913bp, meaning that the samples were positive for Babesia spp. and Theileria spp. respectively (Figure 1). While for the second PCR amplification reaction using single forward primer (Bec-UF2) and two reverse primers, one of them (Cab-R) was specific for B. caballi and another (Equi-R) for T. equi, visualized that the DNA bands size was 392 bp and 540 bp, indicating that the samples were positive for T. equi and B. caballi, respectively (Figure 2).
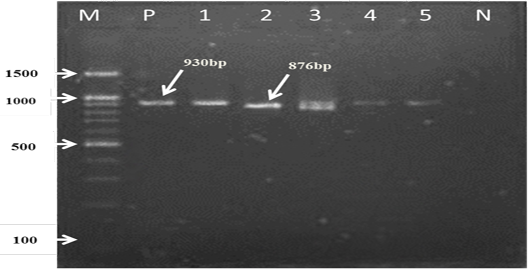
Figure 1: PCR detection of B. caballi and T. equi with a set of primer combinations (Bec-UF1 and Bec-UR): M= 100 bp DNA marker; Lanes 1-2=B. caballi in approximately band size 876bp; lanes 3-6= T. equi in approximately band size 930bp; Lane N) negative control.
Detection rate of T. equi and B. caballi and both protozoa in ixodid ticks using m-PCR
Out of 50 examined adult ticks 10 (20%) were positive for equine piroplasmosis (Table 3). DNA of T. equi was detected in 8 (16%) adult ticks of species H. marginatum marginatum (n=2), H. anatolicum exacavatum (n=1), Rhipocephalus turanicus (n=1), Boophilus microplus (n=1) and
Table 1: The Oligonucleotide primers used to amplify the hemoparasites 18S rRNA genes of piroplasms. (Alhassan et al., 2005).
| Primers | Sequences 5’-3’ | Target gene | Expected size (bp) |
| Bec-UF1 |
5’-GTTGATCCTGCCAGTAGTCA-3’ |
“Catch-all” (Babesia spp. and Theileria spp.) |
876 Babesia spp. |
| Bec-UR |
5’-CGGTATCTGATCGTCTTCGA-3’ |
913 Theileria spp. |
|
| Bec-UF2 |
5’-TCGAAGACGATCAGATACCGTCG-3’ |
Specific primers for B. caballi and T. equi |
------- |
| Cab-R |
5’-CTCGTTCATGATTTAGAATTGCT-3’ |
540 | |
| Equi-R |
5’-TGCCTTAAACTTCCTTGCGAT-3’ |
392 |
Bec-UF1 and Bec-UF2: catch-all forward primers; Bec-UR: catch-all reverse primers; Cab-R: B. caballi-specific reverse primer, Equi-R: T. equi-specific reverse primer.
Table 2: Prevalence and ratio of Ixodid tick according to sex infesting equine in Erbil province.
| Specimens of adult ticks | Male no. (%) | Female no. (%) | Total no. (%) | ♂/♀ |
P value |
| Hyalomma marginatum | 7 | 5 | 12 (12.2%) | 1.45:1 | 0.315 |
| Hyalomma exacavatum | 9* | 1 | 10 (10.2%) | 9.37:1 | 0.103 |
| Boophilus microplus | 2 | 14* | 16 (16.3%) | 0.14:1 | 0.942 |
| Boophilus annulatus | 21 | 18 | 39 (39.8%) | 1.21:1 | 0.001** |
| Rhipocephalus turanicus | 9 | 12 | 21(21.4%) | 0.78:1 | 0.86 |
| Total | 48 | 50 | 98 | 0.96:1 |
*: (p>00.5); **: (p>00.1).
B. annulatus (n=3). DNA of B. caballi was detected in 1 (2%) adult ticks of species Rhipocephalus turanicus (n=1). DNA of both protozoa were detected in 1 (2%) adult ticks of species B. annulatus (n=1). This finding that T. equi was more common detected in ticks compared with B. caballi and both protozoa.
Table 3: Detection of T. equi and B. caballi and both protozoa in Ixodid ticks using m-PCR.
| Type of protozoa | |||||
| Tick species | No. of tested ticks | No. of positive ticks | T. equi | B. caballi | Both protozoa |
| HM | 10 | 2 | 2 | 0 | 0 |
| HE | 10 | 1 | 1 | 0 | 0 |
| RT | 10 | 2 | 1 | 1 | 0 |
| BM | 10 | 1 | 1 | 0 | 0 |
| BA | 10 | 4 | 3 | 0 | 1 |
| Total | 50 | 10 (20%) | 8 (16%) | 1 (2%) | 1 (2%) |
No.: Number of samples; HM: Hyalomma marginatum; HE: Hyaloma exacavatum; RT: Rhipocephalus turanicus; BM: Boophilus microplus; BA: Boophilus annulatus.
Phylogenic analysis
T. equi and B. caballi sequences for the hypervariable V4 regionof the 18SrRNA gene were obtained from four and five samples respectively. The sequences were deposited in GenBank with some subsequently employed for the phylogenic analysis.
These sequences when analysed by BLASTn shared identify ranging from 98-100% with T. equi and B. caballi sequences previously detected. The sequences were clustered in two different groups, B and C for T. equi while for B. caballi they clustered to group B.
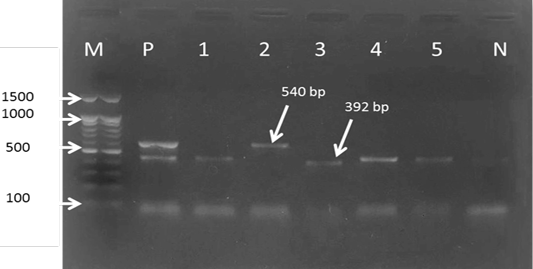
Figure 2: Gel electrophoresis image showing: lane M) 100bp DNA ladder; Lane P) positive control for T. equi and B. caballi; Lane1-4) multiplex PCR detected T. equi and B. caballi in approximately band size 392 and 540 bp respectively; Lane N) negative control.
The tree of T.equi shows two major clades representing the genotype (B and C) Figure 3. Two sequences (MH368310 and MH368318) detected in ticks were clusterd with sequences previously detected from Iran (KF704750), Egypt (MF192854), Jordan (KX227638), Brazil (KJ573337), Cuba (KY111760) and Turkey (MG569905) belong to the clade C, and one sequences (MH368342) were posisioned near to sequences previously detected from Turkey (MG569901), Trindida (KU289006), Sudan (AB515315), Turkey (KU921663), Brazil (MG052902),South Africa (EU642508), Jordan (KX227637), India (LC042536) and Iran (KJ418423) belong to the clade B.
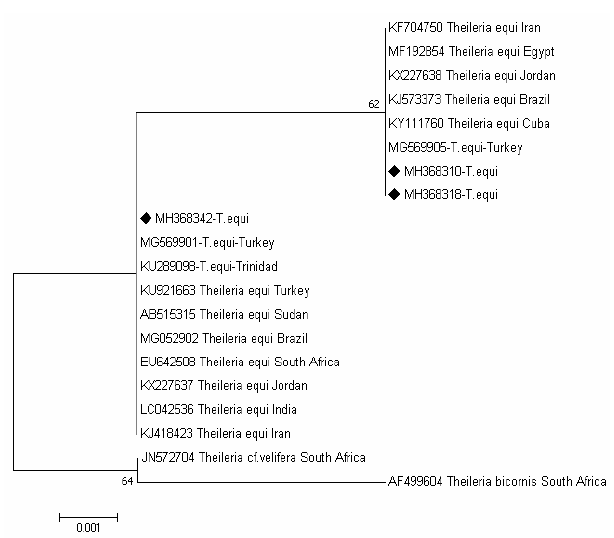
Figure 3: Rooted phylogenic tree of Theileria equi as interfered from partial sequences of 18S rRNA gene. Black diamond indicated sequences obtained from the present study; others represent sequences from references.
Tree of Babesia caballi shows one clades representing the known genotype of Babesia caballi genotype B Figure 4, one sequence (MH368304) were positioned near to the sequences previously detected from Jordan (JN596979), Mongolia (JQ288735), South Africa (EU642513), (EU888904), (Z15104) and (EU888901), Jordan (JQ417262) and Malaysia (KU879026) and (KU879023) belong to the clade B.
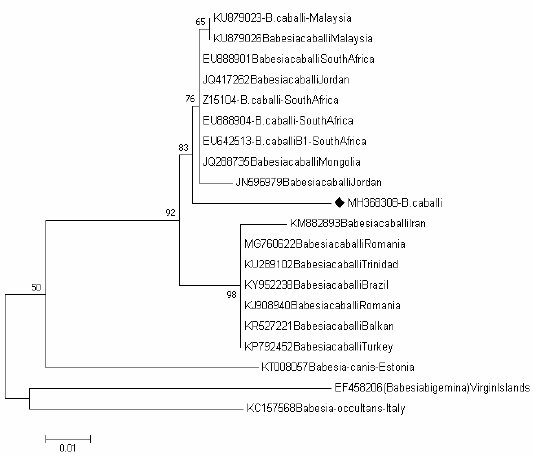
Figure 4: Rooted phylogenic tree of Babesia caballi as interfered from partial sequences of 18S rRNA gene. Black diamond indicated sequences obtained from the present study; others represent sequences from references.
Ticks and tick-borne diseases have a great economical and vital importance throughout the world, and they affect animals health through transmitting of different diseases including: protozoan, bacterial, rickettsial, and viral agents (Ozdarendeli et al., 2008). Equine piroplasmosis which are important tick-borne diseases infect a great variety of equids (Jongejan and Uilenberg, 2004).
Several studies have conducted about tick-borne diseases in equids and other animals using different methods in some regions of Iraq (Estrada-Peña et al., 2004; Alsaad et al., 2010; Aziz et al., 2019). The scarcity of literature related to equids showing any previous molecular study targeting tick population responsible for transmission of theileriosis and babesiosis in Erbil state has not performed any comparison on the prevalence of ixodid tick vectors in the region.
In this study, five ixodid species were identified morphologically on equids during the study, However, according to the (Estrada-Peña et al., 2004). The comparison between these results with other previous studies carried out in different parts of Iraq and other neighboring countries indicated that the main three genera; Hyalomma, Rhipocephalus and Boophilus were observed (Omer et al., 2007; Hasson, 2012; Biglari et al., 2018; Aziz and AL-Barwary, 2019b)
Among Five tick species examined in terms of Theileria equi and Babesia caballi species, all of them were positive for Theileria equi while just one species Rhipocephalus turanicus and Boophilus. annulatus was positive for Babesia caballi and both protozoa respectively.
A previous study conducted in Jordan reported that the prevalence rate of infections was 18.8% for T. equi and 7.3% for B.caballi in horses used PCR technique (Qablan et al., 2013). Another study demonstrated that the infection rate of Equine piroplasmosis was 4.93% in Turkey; 2.96% and 1.97% for T. equi and B. caballi respectively (Kizilarslan et al., 2015).
In the present study, the highest infection rate of Equine Piroplasmosis was found in Boophilus annulatus as compared to other identified ticks. H. anatolicum anatolicum was established in the previous study as the main vector responsible for transmission of EP among the population equine (Kumar et al., 2007). This may be due to the climatic change in last decade and the little number of ticks which observed in horses during this survey due to the great interest in periodic hygiene of horses as a continuous horses cleaning.
The data found that the infected rate of Theileria equi in Ixodid ticks by m-PCR was statistically more significant than that of B. caballi (P<0.05). These findings are agreed with a report by others whom study horses and donkeys from Iraq and neighbouring countries using different techniques (Alsaad et al., 2012; Kizilarslan et al., 2015). This situation is likely to be as result of the effectively elimination of B. caballi by active treatment and host immune system, in contrast to the long-life persistence of T. equi (Laus et al., 2015).
The 18S rRNA gene is widely used for molecular detection and phylogenic analysis of piroplasms in equids (Hall et al., 2013). Here in the molecular detection and phylogenic analysis of T. equi and B. caballi sequence amplified among ticks from Erbil province were assessed and appear to be the first study of its kind from this area.
The present study has provided first data on the genetic diversity of piroplasms in equids population from Iraq based on 18S rRNA gene sequences. Until today, five genotypes of T. equi and three genotypes of B. caballi lineages were established in equine populations from different countries (Bhoora et al., 2010). However by finding a new sequence from various geographical area, further increase in the genetic diversity of T. equi and B. caballi has been predictable (Qablan et al., 2013). This variation in distribution of piroplasms 18S rRNA gene sequences may be related to the horse and mule business movement from other countries especially neighbor countries that primarily included turkey, Iran and Jordan.
The T. equi isolates in the current study clustering within genotypes (B and C), this may reveled an overlapping occurrence of different genotypes within the same population of equids and tick vectors, or may represent a novel variant would require further confirmation (Qablan et al., 2013; Salim et al., 2013).
The B. caballi isolates obtained in our study clustered within the genotype B which corresponds to B. caballi was previously isolated from Jordan (JN596979), Mongolia (JQ288735), South Africa (EU642513), (EU888904), (Z15104) and (EU888901), Jordan (JQ417262), Malaysia (KU879026) and (KU879023).
In results, the high prevalence rate of T. equi was found in ticks in Erbil province, although this considerable diversity in 18SrRNA was found in T. equi, may be related to the history horse business movement between neighboring and our country by illegal modes. The importation of horses into Kurdistan region of Iraq has more involved in the last decade.
CONCLUSIONS AND RECOMMENDATIONS
It has been concluded that five tick species were prevalent from equids in Erbil province, the infected rate of Theileria equi in ticks by conventional PCR was statistically more higher than B. caballi. The molecular diagnostic tool in the detection of T. equi and B. caballi from ticks has been used for the first time in Erbil province in this study. In order to provide better insight of epizootiological situation of tick prevalence and transmission cycle of equine piroplasmosis in this region of our country.
ACKNOWLEDGMENT
We want to thank Dr. Nazhad H. Qader and Dr. Yunis A. Ahmad and the farmers for their assistance with the sample collection. We would also like to thanks Mr. Muhsin J. Abdulwahid (Scientific Research Center/ Salahaddin University) for their help to facilities using laboratory of molecular biology.
Author’s Contribution
All authors contributed equally.
Conflict of interest
The authors have declared no conflict of interest.
References






