Advances in Animal and Veterinary Sciences
Research Article
Genetic Variation of E. coli Strains Isolated from Poultry Slaughterhouses at Ismailia Governorate, Egypt
Mahmoud Ezzat1, Ali Wahdan1*, Fatma Yousef2, Manal Munier2
1Department of Bacteriology, Immunology and Mycology, Faculty of Veterinary Medicine, Suez Canal University, 41522 Ismailia, Egypt; 2Department of Clinical Pathology, Animal Health Research Institute, Ismailia, Egypt.
Abstract | Shiga toxin producing E. coli is represented as one of the main source of foodborne infectious disease distributed all over the world. This area of study not fully explained before at Ismailia governorate. So this study aimed to make genetic survey of all isolated E. coli strains with attention to shiga toxins isolated from different sections of poultry slaughterhouses and workers at Ismailia governorate. One hundred and fifty swab samples were collected from baskets, workers hands, machines, processing tools and food contact surfaces and subjected to bacteriological examination. Ten samples were positive for E. coli with a percentage 6.6%. Different serogroups of E.coli isolated from poultry slaughterhouses were O63:H7 (2), O125:H5, O63:H5, O119:H6, O125:H2, O112ac:H2, O136:H2, O127:H2, O1:H2. Nine E.coli strains were subjected to PCR for genetic detection of Shiga-like toxins genes (stx 1 and stx 2), attaching and effacing (eae A) and enterohaemolysin gene (hly) gene in isolates. 5 E.coli isolates showed positivity for the stx1 gene (55.5%), 2 isolates for stx2 gene (22.2%) and 4 isolates for eaeA gene (44.4%). While, all isolates showed negativity for hly gene. In conclusion, genotypic survey proved the presence of shiga toxins producing E. coli and other virulence E.coli strains at examined slaughterhouses.
Keywords | E. coli, Poultry slaughterhouses, Virulence factors.
Editor | Kuldeep Dhama, Indian Veterinary Research Institute, Uttar Pradesh, India.
Received | July 22, 2018; Accepted | August 19, 2018; Published | October 17, 2018
*Correspondence | Ali Wadhan, Department of Bacteriology, Immunology and Mycology, Faculty of Veterinary Medicine, Suez Canal University, 41522 Ismailia, Egypt; Email: dr_aly_w@yahoo.com
Citation | Ezzat M, Wahdan A, Yousef F, Munier M (2018). Genetic variation of E. coli strains isolated from poultry slaughterhouses at ismailia governorate, egypt. Adv. Anim. Vet. Sci. 6(12): 531-536.
DOI | http://dx.doi.org/10.17582/journal.aavs/2018/6.12.531.536
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2018 Ezzat et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
Poultry meat and its products are very popular food in Egypt as well as throughout the world. The source of contamination may cross to the poultry meat during slaughtering and processing leading to introduction of pathogens into the meat. The source of these pathogens may be coming from the gastrointestinal tract or from the surrounding environment in farm and/ or slaughterhouse. Poultry is the most common food source of human infection byenteric pathogens throughout the world (Samaha et al., 2012)
Because of the relatively high frequency of contamination of poultry with potential pathogens, raw poultry meats are often known to be sources of infections in man (Soares et al., 2018). So there are certain measures must be followed in poultry meat to prevent possible contamination from digestive tracts, during packaging, from utensils and from handlers (Althaus et al., 2017).
The poor hygienic practices lead to contamination in meat by E.coli. Detection of E.coli in food means that there are other harmful bacteria could be detected in the same food (Beutin et al., 2007). Also, detection of E. coli virulence strains in poultry meat lead to infection for slaughterhouse workers and consumers (Xia et al., 2010).
The virulent E.coli strains especially Stx1 and stx2 toxins producing E.coli, have the ability to attach with intestinal epithelium making hemorrhagic colitis and hemolytic – uremic syndrome. Other important virulence factors of STEC are intimin (encoded by eae A gene), a plasmid-encoded enterohaemolysin (ERFAN).
This study aimed to make agenetic survey of all isolated E. coli strains with attention to shiga toxins isolated from different sections of poultry slaughterhouses and workers at Ismailia governorateand to provide practical information for the poultry industry to establish a cleaning frequency protocol during processing.
MATERIALS AND METHODS
Sampling
A total of 150 swab samples were collected under complete aseptic condition from baskets, workers hands, machines, processing tools and food contact surfaces in 3 poultry slaughterhouses at Ismailia governorate. Each sample transported in buffered peptone water and subjected to bacteriological examination.
Isolation and Identification of E. coli
A Loopful from the incubated broth was streaked onto MacConkey s agar and blood agar plates and incubated at 37ºC for 24 h. Lactose fermenting colonies were picked up and streaked onto EMB agar and incubated at 37 ºC for 24 h. Metallic green sheen colored colonies on EMB. Suspected E. coli isolates were identified morphologically by Gram stain, motility test and biochemically according to (Quinn, Markey et al., 2011).
Serotyping of E. coli Isolates
The somatic (O) and Flagellar (H) antigens were carried out according to (Cowan and Steel, 2004). Serotyping was carried out at the serology department in Animal Health Research Institute in Dokki, Cairo, Egypt.
Genetic Detection of Isolated E.coli Virulence Genes
Nine E. coli strains were subjected to PCR for detection of shiga toxins (stx 1 and stx 2), eae A and hly genes. QIAamp DNA Mini Kit used for extraction of DNA . Primers used were shown in Table 1. Temperature and time conditions of the primers during PCR are shown in Table 2. Amplified PCR product was analyzed by electrophoresis on a 1.5% agarose gel stained with 0.5 µg of ethedium bromide/mL. The gels were photographed by a gel documentation system.
Table 1: Oligonucleotide primers of E.coli virulence genes
| Reference | Length of amplified product | Primer sequence (5'-3') | Target gene |
|
(Dipineto, Santaniello et al. 2006)
|
614 bp | ACACTGGATGATCTCAGTGG | Stx1 |
| CTGAATCCCCCTCCATTATG | |||
| 779 bp | CCATGACAACGGACAGCAGTT | Stx2 | |
| CCTGTCAACTGAGCAGCACTTTG | |||
| (Piva, Pereira et al. 2003) | 1177 bp | AACAAGGATAAGCACTGTTCTGGCT | Hly |
| ACCATATAAGCGGTCATTCCCGTCA | |||
| (Bisi-Johnson, Obi et al. 2011) | 248 bp | ATGCTTAGTGCTGGTTTAGG | eaeA |
| GCCTTCATCATTTCGCTTTC |
Table 2: Temperature and time conditions of the used primers during PCR.
| Final extension | No. of cycles | Extension | Annealing | Secondary denaturation | Primary denaturation | Target gene |
|
|
72˚C 10 min. |
35 |
72˚C 45 sec. |
58˚C 45 sec. |
94˚C 30 sec. |
94˚C 5 min. |
Stx1 and stx2 |
|
|
72˚C 10 min. |
35 |
72˚C 1 min. |
60˚C 50 sec. |
94˚C 30 sec. |
94˚C 5 min. |
Hly |
|
|
72˚C 7 min. |
35 |
72˚C 30 sec. |
51˚C 30 sec. |
94˚C 30 sec. |
94˚C 5 min. |
eaeA |
|
RESULTS
The bacteriological examination of 150 Swab samples (50 swab samples from different stations of each poultry slaughterhouse) revealed in 10 samples (6.6%) positive reaction to E.coli as shown in the Table 3. All isolates showed the typical colonies characteristic of E.coli where it appeared a smooth, shiny and strong lactose fermenting (pink) colonies on MacConkey agarand metallic sheen green colonies on Eosin Methylene Blue. On blood agar, colonies were non hemolytic, gray, white, moist, glistening, opaque, circular and convex with entire edges.
Serotyping of E.coli isolates recovered from poultry slaughterhouses: The isolated E.coli strains were serogrouped in to O1, O63, O112, O119, O125, O127 and O136. Untypable strains were also recovered shown in Table 4.
Table 3: Prevalence of E.coli isolates from different stations of poultry slaughterhouses swab samples
|
Samples |
Number of Swab samples |
E.coli isolates |
|||
| Positive | Negative | ||||
| No. | % | No. | % | ||
|
Poultry slaughterhouse (A) dirty area clean area |
25 25 |
1 0 |
4 0 |
24 25 |
96 100 |
|
Poultry slaughterhouse (B) dirty area clean area |
25 25 |
1 0 |
4 0 |
24 25 |
96 100 |
|
Poultry slaughterhouse (c) dirty area clean area |
25 25 |
5 3 |
20 12 |
20 22 |
80 88 |
Table 4: Prevalence of E.coli serotypes from different poultry slaughter-houses
| Number of sample | Serology | Farm | Sample station |
| (2) | O63:H7 | (C) | Dirty area"machine" |
| (1) | O125:H5 | (A) | Dirty area"machine" |
| (1) | O63:H5 | (B) | Dirty area"machine" |
| (1) | O119:H6 | (C) | Dirty area"machine" |
| (1) | O125:H2 | (C) | Dirty area"machine" |
| (1) | O112ac:H2 | (C) | Clean area(food contact surface"liver" ) |
| (1) | O136:H2 | (C) | Clean area (Worker hand) |
| (1) | O127:H2 | (C) | Dirty area"machine" |
| (1) | O1:H2 | (c) |
Clean area(worker hand) |
According to genetic characterization of virulence genes (stx1, stx2, eaeA, hly) by PCR. 5 E.coli isolates showed positivity for the stx1 gene (55.5%), 2 isolates for stx2 gene (22.2%) and 4 isolates for eaeA gene (44.4%), as shown in Table 5 and Figures 1, 2, 3.
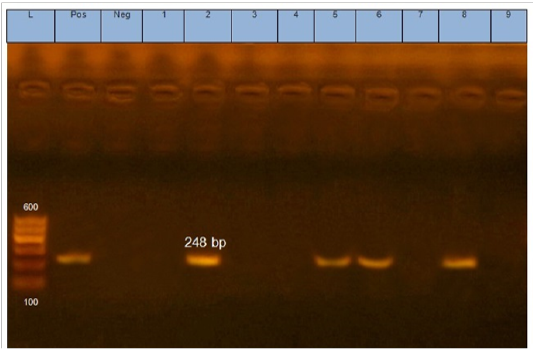
Figure 1: Gel electrophoresis of amplified eaeA gene (248bp): L= molecular weight marker (100bp), 2,5,6 and 8 E.coli samples were positive for eaeA gene. Neg=control negative reaction .pos=positive (previously known eaeA positive E. coli).
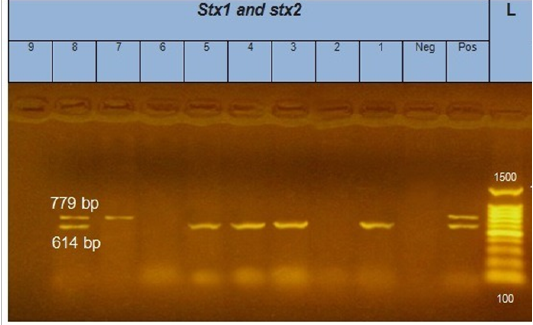
Figure 2: Gel electrophoresis of amplified stx1, stx2 genes (614bp), (779bp) respectively : L= molecular weight marker (100bp)=1,3,4,5 and 8 E. coli samples were positive for stx1 gene. While, 7 and 8 E. coli samples positive for stx2 gene. Neg= control negative reaction .pos=positive (previously known stx1, stx2 positive E. coli).
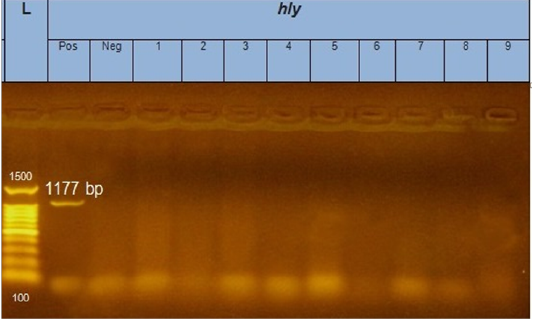
Figure 3: Gel electrophoresis of amplified hly gene (1177bp): L= molecular weight marker (100bp), from 1 to 9 E.coli samples were negative for hly gene. Neg=control negative reaction .pos=positive (previously known hly positive E.coli).
Table 5: Detection of virulence genes in E.coli isolates by PCR
| Serotype | eaeA | stx1 | stx2 | hly | Sample position |
|
O63:H7 |
- | + | - | - | (C) Dirty area machine |
| O125:H5 | + | - | - | - | (A) Dirty area machine |
| O63:H5 | - | + | - | - | (B) Dirty area machine |
| O119:H6 | - | + | - | - | (C) Dirty area machine |
| O125:H2 | + | + | - | - | (C) Dirty area machine |
| O112ac:H2 | + | - | - | - | (C) clean area (food contact surface"liver") |
| O136:H2 | - | - | + | - | (C) clean area worker hand |
| O127:H2 | + | + | + | - | (C) Dirty area machine |
| O1:H2 | - | - | - | - | (C) Clean area worker hand |
DISCUSSION
The bacteriological examination of 150 Swab samples (50 swab samples from different stations of each poultry slaughterhouse) revealed in 10 samples (6.6%) positive reaction to E. coli as shown in the Table 3. Seven samples (70%) from dirty area including machines, workers hands and carcasses and 3 samples (30%) from clean area including machines, workers hands and product. As in dirty area workers hand, evisceration machine, scalding tank, act as a source of E.coli contamination (Van den Bogaard et al., 2001; Farah et al., 2007) Who found the E. coli counts were around 3.0 log CFU/cm2 at the scalding machine. Generally, the presence of E. coli in poultry slaughterhouse considered as an indicator for improper handling or unhygienic conditions, which agreed with (Samaha et al., 2012).
The negativity of all the isolates to hemolysis on 5% sheep blood agar was in accordance with (Peer et al., 2013) who attributed heavy mortality in chicks due to non –hemolytic strains indicating that pathogenic E.coli to be independent of hemolytic activity and (Literak et al., 2013) who reported that none of their isolates was positive for hemolysis on 5% sheep blood agar. However, the hlyA gene was not detected in strains tested negative for enterohemolysin that was differ with (Farhan et al., 2014) who detect hlyA in seven strains tested negative for enterohemolysin.
The serogrouping of isolated E.coli strains from examining sample swabs were O63:H7 (2), O125:H5, O63:H5, O119:H6, O125:H2, O112ac:H2, O136:H2 O127:H2, O1:H2 as shown in Table 4, is nearly similar to the obtained results by (Rey et al., 2006), as E. coli O125, O86 and O119 were previously isolated from chicken meat in Egypt. Also, (Farah et al., 2007) identified E. coli from chicken; O27, O78 and O86 while (Oh et al., 2012) recorded the predominance of E. coli serotype O125 (7 isolates), followed by O142, O124, untypable (3 isolates, each), O126, O78 (2 isolates, each), O25, O127, O91, O86, O119 (one isolate, each) isolated from poultry slaughterhouse. While, in chilled chicken carcasses by (Nossair et al. , 2015) recorded O26 by 16%, O86 by 16%, O119 by 12%, O126 by 24% and O124 by 8% and in freshly slaughtered chicken carcasses, were isolated O26 by 18%, O86 by 16%, O119 by 14%, O126 by 14% and O124 by 18%.
In the present study, 5 E.coli isolates showed positivity for the stx1 gene, with an incidence of 55.5%, 2 isolates showed positivity for stx2 gene, with incidence of 22.2%, 4 isolates showed positivity for eaeA gene, with incidence of 44.4%, all isolates showed negativity for hly gene. Regarding to the occurrence of stx1 gene in 9 E.coli strains, only 5 E.coli strains were detected, containing stx1 gene by percent of 55.5%.This result was different from finding which was recorded by (Momtaz and Jamshidi, 2013) who showed that 100% of EHEC serogroups were positive for all stx1.
Regarding to the occurrence of stx2 gene in 9 E.coli strains, only 2 E.coli strains were detected, containing stx2 gene by percent of 22.2%.This result was nearly similar to finding which was recorded by (El-Jakee et al., 2012) who detected stx2 by percentage 41.67% in 5 E.coli isolates out of 12 one.
Other studies, documented in the absence of both types of Shiga toxin type 1 and type 2 genes in E.coli isolates from various types of samples (Fereidouni et al., 2009; Murthy and Rao 2014).
Concerning to examination of E.coli strains for the presence of Intimin gene, results detected 4 out of 9 E.coli strains, by percent of 44.4%. These findings were nearly agreed with those obtained from (Dutta et al., 2011) who detected eaeA in 4 E.coli strains out of 10 samples by percent of 40% and (El-Jakee et al., 2012) who detected eaeA in 5 E.coli strains out of 12 by percentage 41.67%.
In this study all 9 E.coli isolates showed negativity for hly gene, the present findings do not coincide with what was reported by (Dutta et al., 2011) who detected hly in 7 E.coli strains out of 10 samples by percentage 70%. Meanwhile, the shiga toxin positive isolates were negative by PCR for the eaeA and hly genes were reported by (Parreira and Gyles, 2002; Zakeri and Kashefi, 2012). Our results confirmed the presence of virulence genes including stx, eaeA. Therefore, the environment, tools, machines and handlers of slaughterhouses may be a reservoir of the organism that can cause serious diseases in humans. These results occurred by (Sirotkin et al., 2006) who reported that Shiga toxin-producing E.coli (STEC) has been demonstrated to exist in E.coli samples which is considered as a threat for human health.
PCR is a powerful molecular biology technique for the detection of virulence genes. It is not only highly sensitive and specific, but it also provides rapid and reliable results. Detection of virulence genes was determined in the present study as a molecular tool for identification of E.coli isolates using the PCR technique.
CONCLUSION
Protection of poultry meat from exposure to different types of bacteria during the manufacturing stages is to protect the national economy as well as the protection of human health. stx1, stx2 and eaeA virulence genes were detected examined slaughterhouses at Ismailia, Egypt. So applying of strict hygienic measures inside poultry slaughterhouses urgent needs to decrease the opportunity for access of food poisoning microorganisms. A comprehensive educational program about the potential hazards associated with bacterial infection in foods of animal origin should be in place.
ACKNOWLEDGEMENTS
Authors wish to thank Animal Health and Research Institute (AHRI) for the kind aid in serological and molecular investigations.
CONFLICT OF INTEREST
No conflicts of interests are announced by authors.
Authors Contribution
All authors contributed in writing of this research article. Both FatMa YousEF and Manal MuniEr contributed in collection of samples and traditional identification of E. Coli. While, MahMoud Ezzat and ali Wahdan contributed in serological and molecular characterization of isolated strains.
References






