Advances in Animal and Veterinary Sciences
Research Article
Canine Parvo Enteritis Infection in Egypt: Isolation, Molecular Characterization and Sequencing
Ali A. Abdel-Rhman1*, Farouk A. El Balkemy2, Nasser Z. Abouzeid2, Samir M. Edries3
1Veterinarian in Prison Sector Ministry of Interior; 2Animal Medicine Department, Infectious Diseases, Faculty of Veterinary Medicine, Zagazig University, 44511, Egypt; 3Veterinary Serum and Vaccine Research Institute, Abassia, Cairo.
Abstract | Regular updating of our knowledge regarding canine parvovirus infection (CPV) diagnosis is crucial to increase the awareness of disease significance and subsequently to improve vaccination programs. This work aimed for isolation, identification, molecular characterization and sequencing of the CPV strain of naturally infected dogs in Egypt. A total of 120 dogs of different ages and breeds suffering from vomiting, bloody diarrhea, dehydration, and ruffled hair were clinically examined. The Rapid Immune Chromatographic Test (IC) and culturing on Vero cell were used to test fecal samples (n=120). The findings revealed that (69 and 48/ 120) samples (57.5 and 40 %) were positive for CPV using IC test and CPV’s specific cytopathic effect on Vero cell respectively. The virus identified by Rapid Slide Agglutination (RAT), Virus Neutralization Test (VNT) and Direct Fluorescent Antibody Technique (FAT). Molecular characterization was performed by using the Hfor/Hrev primers for 3 positive samples by IC alone and 3 positive samples by both IC and cell culturing in addition to 6 negative samples. The positive samples were detected at 630 bp for CPV and 427 bp for CPV-2a. Three molecularly positive samples were genotyped using specific primers for CPV-2a, CPV-2b, and CPV-2c. Current study findings revealed that the three sequenced samples are strongly close to the CPV type 2a. We could conclude that CPV-2a is the current type of CPV circulating in Egyptian dog’s population.
Keywords | Canine parvovirus, Egypt, Enteritis, Molecular characterization, Sequencing
Received | September 12, 2019; Accepted | October 26, 2019; Published | December 19, 2019
*Correspondence | Ali A. Abdel-Rhman, Veterinarian in Prison Sector Ministry of Interior; Email: dr_alikabir@yahoo.com
Citation | Abdel-Rhman AA, El Balkemy FA, Abouzeid NZ, Edries SM (2019). Canine parvo enteritis infection in Egypt: Isolation, molecular characterization and sequencing. Adv. Anim. Vet. Sci. 7(s2): 117-122.
DOI | http://dx.doi.org/10.17582/journal.aavs/2019/7.s2.117.122
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2019 Abdel-Rhman et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
Canine parvovirus enteritis in young dogs remain one of the most important cause of morbidity and mortality, and the incidence of the causative virus is continued partly due to ability of the virus to “reinvent” itself and advance into new resistant and more virulent strains (Goddard and Leiswitz, 2010). Canine parvovirus (CPV) belongs to the family Parvoviridae and is closely related to the mink enteritis virus and feline panleukopenia virus (Parrish et al., 1985). CPV is a single-stranded DNA virus that is surrounded by a protein shell (Carmichael and Joubert, 1983; Rimmelzwaan et al., 1990). CPV-2 was rapidly replaced by a new antigenic, termed CPV-2a variant by changing genetically and antigenically, in 1978 that reclaimed the capability to infect felids due to five or six amino acid substitutions located in the VP2 gene (Parrish et al., 1985). Later, new antigenic variations in 1984 in the United States and 2000 in Italy were called (CPV-2b) or (CPV-2c) (Parrish et al., 1985, 1991; Buonavoglia et al., 2001). CPV infection has two characterized prominent clinical forms regarding the age; first one includes dogs of all ages and infected one has enteritis that leads to vomiting and dehydration; while in second clinical form, puppies having an age of less than 3 months are susceptible to CPV. Infected puppies have myocarditis that leads to heart failure (Woods et al., 1980).
Diagnosis of CPV-2 infection is very important as diarrhea in dogs can be developed by several other pathogenic organisms making the diagnosis difficult. Therefore, several serological and molecular diagnostic assays have been developed over the years for rapid, accurate and sensitive diagnosis (Buonavoglia et al., 2001). Electron microscopy is one of the several laboratory methods and have been advanced to identify CPV-2 in the feces of diseased dogs (Amo et al., 1990), conventional and real-time PCR using for viral identification (Desario et al., 2005). This work aimed for isolation, identification, molecular characterization and sequencing of the CPV strains of recently recorded cases in dogs in Egypt.
MATERIALS AND METHODS
This study has been conducted following the guidelines of the Zagazig University Institutional Animal Care and Use Committee, approval number ZU-IACUC/3/S/29/2018.
Animals and sampling
A total of 120 dogs of different breeds and ages suffering from fever, vomiting, bloody diarrhea, dehydration and ruffled hair either vaccinated or non-vaccinated were clinically examined according to Cote (2011). During January 2017 to January 2019, fecal swabs were collected from suspected dogs (n=120) from private pets clinics in Cairo and from educational clinics, Faculty of Veterinary Medicine, University of Zagazig, Egypt. The swabs put in labeled tubes containing normal saline with 10% of antibiotic stock solution (1gm streptomycin sulfate and 1 million IU of penicillin-G sodium I 100 ml saline) and transferred to the laboratory of (Department of Pet Animal Vaccine Research (DPAVR); Veterinary Serum and Vaccine Research Institute (VSVRI)). The samples subjected to 2 freezing and thawing cycles and then centrifuged at 2000 rpm for 10 minutes. The supernatant fluid was separated and kept in -80 ºC till used for virus isolation (Hendrix and Robinson, 2012).
Screening and detection of viral antigen by rapid chromatographic slide immunoassay
Rapid CPV Ag test kit used for the detection of CPV antigen in collecting fecal samples following the manufacturer’s instructions (BioNote, Inc., Gyeonggi-do, Korea).
Virus isolation
CPV-2a isolation was performed in Abassia, Cairo, Egypt, Department of Pet Animal Vaccine Research (DPAVR); Veterinary Serum and Vaccine Research Institute (VSVRI). Isolation of CPV-2a was done on confluent monolayer cell culture of African green monkey kidney cell line (Vero) using a minimum essential medium (MEM) with newborn calf serum (10% for growth medium and 2% for maintenance medium) through 3 blind successive virus passages according to Hirayama et al. (2005).
Virus identification
Normal and infected cell cultures prepared on coverslips were stained with hematoxylin and eosin according to Carleton (1967) to demonstrate the induced CPE of CPV. The presumed CPV isolates were subjected to virus neutralization test using specific anti-CPV serum according to Rossiter and Jessette (1982) the test included cell and virus controls. Daily microscopic examination for detection of any CPE in inoculated cell cultures. Also, direct FAT was carried out on infected Vero cell culture according to Florence et al. (1992).
DNA extraction and cycling conditions of the primers during PCR
It was carried out following QIAamp DNA mini kit instructions. Temperature and time conditions of the different primers supplied by Metabion (Germany) and have specific sequence and amplify a specific product during PCR according to Buonavoglia et al. (2001) (Table 1).
Sequencing of CPV and phylogenetic analysis
Sequence analysis was performed using version 1.83 of the MegAlign module from the CLUSTAL W multiple sequence alignment program, Lasergene DNAStar software Pairwise, which was designed by Thompson et al. (1994). In MEGA6, phylogenetic analysis was performed using the highest probability, neighbor-joining and maximum parsimony (Tamura et al., 2013).
Statistical analysis
Microsoft Excel program was used to analyze all information collected through history, basic clinical examination, laboratory examinations and result measures coded. Information was at that point imported into Statistical Package for the Social Sciences (IBM SPSS Statistics for Windows, Version 20.0. Armonk, NY: IBM Corp). Concurring to the sort of data subjective represent as number and percentage, quantitative proceeds group represent by mean ± SD, the following tests were utilized to test differences for significance, difference, and association of qualitative variable by Chi-square test (c2), agreement by Kappa. P-value was set at <0.05 and <0.001 for significant and high significant level results, respectively.
RESULTS
Rapid chromatographic slide immunassay
Clinically the infected dogs suffered from fever, vomiting, bloody diarrhea, dehydration and ruffled hair (Figure 1).
Rapid CPV test kit was performed on fecal samples of clinically infected dogs (n=120) suffered from fever, vomiting, bloody diarrhea, dehydration, and ruffled hair.
Table 1: Oligonucleotide primers sequences.
| Virus | Gene | Primer/ probe sequence 5'-3' | Length of amplified product (bp) | References |
| Canine parvovirus | VP2 | Hfor: CAGGTGATGAATTTGCTACA | 630 | Buonavoglia et al., 2001 |
| Hrev: CATTTGGATAAACTGGTGGT | ||||
| Pbs: CTTTAACCTTCCTGTAACAG | 427 | |||
| Pbas: CATAGTTAAATTGGTTATCTAC |

Figure 1: Puppies suffering from CPV infection, a. Weakness, dullness, and diarrhea. b. Weakness, dullness, vomiting and bloody diarrhea. c. Weakness, dullness and bloody diarrhea
The test revealed that (69/120) 57.5% of the examined dogs samples were positive for CPV antigen, clear positive line was determined as shown in (Figure 2).
Virus isolation and demonstration of cpv cytopathic effect
Out of 120 samples, 48 samples (40 %) showed progressive cytopathic effect (CPE) with increased virus titer through three consecutive passages of fecal samples. At first, the CPE was initiated by the 5th-day post-infection of Vero cell culture and completed within the 7th to 8th day later then began to appear by the 3rd-day post cell infection and completed by the 7th day. Such CPE was characterized by cell rounding and aggregation followed by cell lysis and detachment from the culture surface (Figure 3b). The virus titer started by 2 log 10 TCID50/ml to reach 5 log 10 TCID50/ml by the third passage.
Virus identification
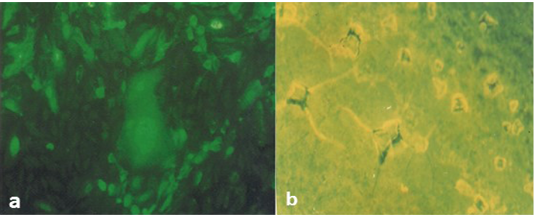
It was found that Rapid slide agglutination (RAT) was able to detect CPV antigens within 2-5 minutes. RAT carried out on Vero cell culture-positive samples at the third passage showed that the strength of agglutination was strong with high titers of virus antigen and weak with lower titers. Although VNT needs 3-5 days to confirm the results of the RAT and the described CPE indicating that the obtained isolate is CPV. Application of direct FAT on infected Vero cell culture using specific anti-CPV serum conjugated with FITC confirming the existence of CPV due to the presence of positive intra-cytoplasmic apple green reaction (Figure 4a).
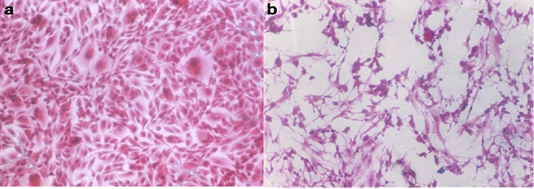
Figure 3: a. Normal Vero cell culture. b. Vero cell culture infected with the obtained CPV showing distorted cell morphology and detached cells (H&E; 100Xs)
Molecular detection and characterization of CPV2
Three positive IC samples were subjected to PCR in addition to three positive samples by both IC and TC along with six negative samples. The six positive samples were detected at 630 bp for CPV (Figure 5a) and 427 bp for CPV-2a (Figure 5b).
Sequencing and phylogenetic analysis revealed that CPV-2a isolates was found in the same cluster with the CPV-2a isolates “KP715709.1”; KP715701.1”; KP715700.1” and “KP715694.1” Phylogenetic investigations of our Egyptian, partial sequences of CPV-2a VP2 recognized monophyletic connections between the closely geographical related CPV strains (Figure 6). GenBank accession numbers for CPV nucleotide sequences were MK614452, MK614453 and MK614454.
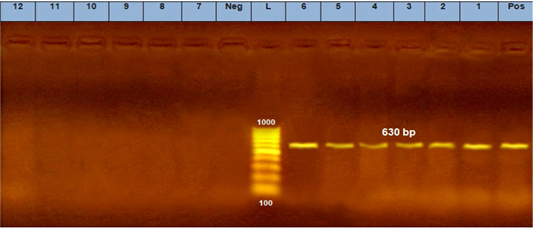
Figure 5: A. PCR assay using CPV-2 primers. (L) DNA ladder 100 bp, lanes (1: 6) positive samples and specific bands at 630 bp. Lane (Pos) : positive control. Lanes (7:12) negative samples, lane (neg) negative control sample.
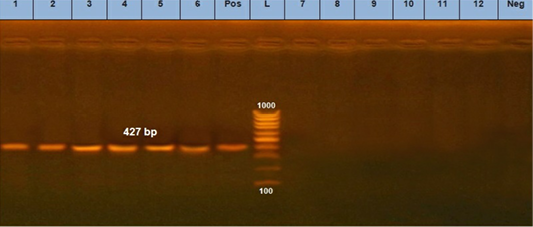
Figure 5: B. PCR assay using using primer set CPV- 2a. (L) DNA ladder 100 bp, lanes (1: 6) positive samples and specific bands at 427 bp. Lane (Pos): positive control. Lanes (7:12) negative samples, lane (neg) negative control sample.
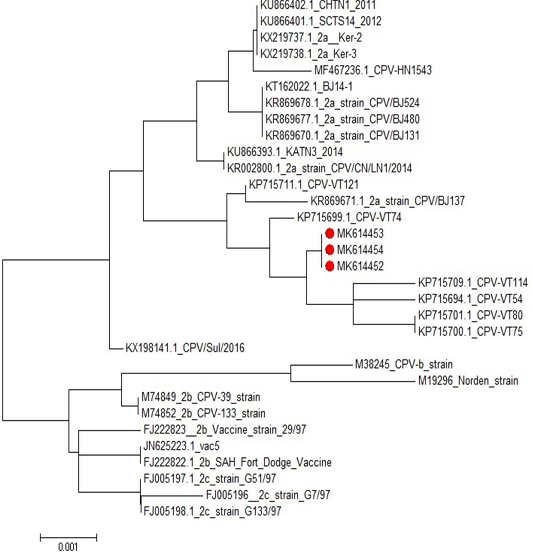
Figure 6: Phylogenetic tree of CPV isolates in this study compared to other sequences in the gene bank.
DISCUSSION
Clinically the infected dogs experienced fever, vomiting, bloody diarrhea, dehydration and ruffled hair. These results were agreed with those mentioned by Saho et al. (2007) and Vasantha (2011) who indicated that bloody diarrhea was one of the most obviously observed signs in 40% of CPV infected animals. As proposed by Bastan et al. (2013), such diarrhea may be due to damage and destruction of the epithelium of intestinal crypts and subsequent villous atrophy. Dehydration was also one of the distinctive symptoms in infected dogs, which is a consequence of a large quantity of fluid loss from vomiting and diarrhea and this was also in agreement with Greene and Decaro (2012). Vomiting, one of the main clinical sign in infected dogs was reported to develop rapidly in 24 to 48 hours after infection as recorded in previous studies (Prittie, 2004; Willard, 2005; Decaro et al., 2005c).
Table 2: Significant association between TC and IC findings.
| Tissue culture | Total |
X2 |
P | Kappa agreement | ||||
| +VE | -VE | |||||||
| IC | +VE | N | 36 | 33 | 69 | 10.02 | 0.0015* | 0.62 |
| % | 75.0% | 45.8% | 57.5% | |||||
| -VE | N | 12 | 39 | 51 | ||||
| % | 25.0% | 54.2% | 42.5% | |||||
| Total | N | 48 | 72 | 120 | ||||
| % | 100.0% | 100.0% | 100.0% | |||||
Rapid CPV (IC) test, applied on fecal samples of diseased dogs revealed that 69 / 120 fecal samples (57.5%) showed positive results (Figure 2). As depicted in Table 2 the IC showed significant association with the result of TC, revealed that True Positive results were 36 samples (75.0%), false-positive results were 33 samples (45.8%), True Negative results were 39 samples (54.2%), false-negative results were 12 samples (25.0%). The validity of (IC) test regarding tissue culture was average as sensitivity was 75%, specificity was 54.2%, +VE Predictive was 52.1%, -VE Predictive was 76.4%, and the accuracy was 62.5% with Kappa agreement 0.62 (Table 3). These results are in line with previous studies (Proksch and Hartmann, 2015), who reported that variation of sensitivity and high numbers of false-negative results are common, which potentially leads to misdiagnosis. Soliman (2018) used rapid antigen test kit, for detection of CPV in fecal samples obtained from naturally infected dogs and found 50% positive samples. Also, our results confirmed by Desario et al. (2005) who stated that immunochromatographic (IC) test sensitivity did not exceed 50% concerning the nucleic acid-based methods, whereas the specificity was 100%. The poor sensitivity of the IC test was associated with the low amounts of virus shed in the feces during the late stages of infection and/or the early presence of high CPV antibody titers in the gut lumen that may sequestrate most viral particles.
Table 3: Validity of IC regards Tc.
| Sensitivity | Specificity | +VE Predictive | - VE Predictive | Accuracy |
| 75% | 54.2% | 52.1% | 76.4% | 62.5% |
In present study, we also observed that 40% (48/120) samples showed progressive CPE on Vero cell culture. It was found that the demonstrated CPE was not so prominent within the 1st passage in the form of increased granularity, distorted cell morphology and detached cells as shown in Figure 3. These findings coming into agreement with that recorded in previous studies (Kotb, 1994; Sutton et al., 2013; Soliman, 2018; El-Gendy, 2018).
Rapid slid agglutination test (RAT) was able to detect the antigens of CPV within 2-5 minutes. RAT carried out on Vero cell culture-positive samples at the third passage showed that the strength of agglutination was strong with high titers of virus antigen and weak with lower titers. This finding also in line with with Soliman (2018). This test due to its simplicity and rapidity of performance and its low cost has a great potential for use for direct detection and identification of CPV as a screening strategy.
Virus neutralization tests indicated the presence of CPV, in this respect, VNT is considered the queen test where it is highly specific and accurate, although it requires cell culture ability, appropriate and expert personnel furthermore the permissive cell lines to be utilized. It has been shown in common and experimental infections that CPV-2 is measurable by viral isolation just for a few days post-infection (Decaro et al., 2005). Desario et al. (2005) and Soliman (2018) confirmed their CPV isolates through the application of VNT. The direct fluorescent antibody technique confirmed the incidence of CPV in Vero cell culture using specific anti-CPV serum conjugated with FITC that revealed positive intra-cytoplasmic apple green reaction (Figure 4a). These results agreed with Kotb (1994) and Soliman (2018) who stated that specific intracellular fluorescence detected in infected cell culture is indicator of CPV presence in infected cell lines.
The diagnosis of CPV infection using PCR technique is a popular technique now a days (Kumar et al., 2010). Our results showed that 6 randomly selected cell culture passage samples showed clearly positive results for parvovirus using Hfor/Hrev primers yielding a specific product size of 427 bp (Figure 5). Additionally, three of such positive samples were subjected to genotyping using specific primers for CPV2a, CPV2b, and CPV2c. The results showed that the three samples were positive for CPV2-a as shown in (Figure 6).
Sequencing and phylogenetic analysis revealed that the presently obtained CPV-2a isolate was found in the same cluster with the CPV-2a isolate KP715709.1; KP715701.1; KP715700.1 and KP715694.1 Phylogenetic investigations of our Egyptian, partial sequences of CPV-2a VP2 recognized monophyletic connections between the closely geographical related CPV samples and GenBank accession numbers for our canine-parvovirus nucleotide sequences were MK614452, MK614453, and MK614454. Furthermore, one of the most frequently used CPV vaccine strains was phylogenetically correlated with partial CPV2a VP2 sequences. Our findings also revealed that the three sequenced samples are strongly close to the genotype of CPV-2a compared to other sequences in the gene bank. CPV-2a and 2b genotypes were detected in Egypt (Morcos, 2013; Fouad, 2014; Soliman, 2018). El-Gendy (2018) also found that recent field CPV strains were 99% homologous to Egypt’s local vaccine strain.
CONCLUSION
Detection of CPV antigen by rapid (IC) test and Rapid slide agglutination test can rapidly diagnose CPV infection. Virus neutralization test along with a fluorescent antibody technique can be employed successfully for confirmatory identification of CPV. The circulating type of CPV in dogs in Egypt is CPV-2a.
ACKNOWLEDGMENTS
This work was done without funding from any organization. Necessary facilities of the Department of Animal Medicine, Faculty of Veterinary Medicine, Zagazig University, Department of Pet Animal Vaccine Research (DPAVR); Veterinary Serum and Vaccine Research Institute (VSVRI) Abassia, Cairo, Egypt were used.
Authors Contribution
Ali A. Abdel-Rhman, Farouk A. El Balkemy, Nasser Z. Abouzeid and Samir M. Edries contributed to the design and implementation of the research, to the analysis of the results and to the writing of the manuscript.
CONFLICT OF INTEREST
The authors have no conflict of interest to declare.
REFERENCES