Advances in Animal and Veterinary Sciences
Research Article
Outer Membrane Protein A Gene Based Diagnosis of Riemerella anatipestifer Infections in Ducks
Ciby Somu, Uma Radhakrishnan*, Shonima Pala
Department of Veterinary Biochemistry, College of Veterinary and Animal Sciences, Mannuthy, Thrissur 680651, Kerala Veterinary and Animal Sciences University, Kerala, India.
Abstract | New Duck disease or infectious serositis or anatipestifer syndrome, an epizootic infectious disease of poultry, caused by Riemerella anatipestifer is responsible for severe mortality in ducks world-wide. The present study reveals the molecular characterization of outer membrane protein A gene (ompA) of R. anatipestifer isolated from ducks of Kerala as well as its application for developing control and diagnostic strategies for New Duck disease. Outer membrane protein A gene of three isolates of R. anatipestifer from ducks of Kerala were amplified by PCR and cloned in T/A vector. After sequencing of the amplicons, the sequence similarity analysis was done using selected software. The sequence analysis of the gene showed that no much variation of ompA existed among the isolates and that all the isolates were similar to a Taiwanese strain of R. anatipestifer. The results emphasise that the ompA gene of R. anatipestifer could be utilised for developing diagnostic and control strategies for New Duck disease.
Keywords | Outer membrane protein, ompA gene, PCR, Riemerella anatipestifer, New Duck disease
Editor | Kuldeep Dhama, Indian Veterinary Research Institute, Uttar Pradesh, India.
Received | September 09, 2016; Accepted | September 19, 2016; Published | October 10, 2016
*Correspondence | Uma Radhakrishnan, Department of Veterinary Biochemistry, College of Veterinary and Animal Sciences, Mannuthy, Thrissur 680651, Kerala Veterinary and Animal Sciences University, Kerala, India; Email: uma@kvasu.ac.in
Citation | Somu C, Radhakrishnan U, Pala S (2016). Outer membrane protein A gene based diagnosis of Riemerella anatipestifer infections in ducks. Adv. Anim. Vet. Sci. 4(10): 527-535.
DOI | Http://dx.doi.org/10.14737/journal.aavs/2016/4.10.527.535
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2016 Somu et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
New Duck disease or Anatipestifer syndrome, an epizootic infectious disease infecting domestic poultry, mainly ducks, is caused by Riemerella anatipestifer, a gram-negative, ubiquitous, nonmotile, non-spore-forming, rod shaped bacterium (Harry, 1969; Sandhu, 2008). The bipolar nature of the organism as well as clinical signs of the disease that simulates Escherichia coli infections and salmonellosis often leads to diagnostic errors. More than 21 serotypes of the organism have been identified by slide and tube agglutination tests with antisera (Pathanosophon et al., 1995; Tsai et al., 2005). The unscrupulous use of antibiotics to control the disease has fastened the emergence of drug resistant strains (Chen et al., 2010, 2012). Such divergence in genetic make-up results in limited cross protection between different R. anatipestifer strains and variations in virulence factors leading to mixed infection with multiple serotypes of the organism in the same bird (Subramaniam et al., 2000; Huang et al., 2002; Yu et al., 2008). Virulence and pathogenicity as assessed by morbidity and mortality rates varies with the serotype involved (Subramaniam et al., 2000). The molecular basis for these differences is not much known, since very few virulence factors of R. anatipestifer could be identified yet.
Outer membrane proteins of pathogenic bacteria generally play an important role in virulence and immunogenicity. Outer membrane protein A (ompA); that helps in maintenance of the structural integrity of cell envelope, bacterial conjugation, bacterial attachment, colicin upake and in porin activity; has been proven to be a virulence factor of R. anatipestifer (Subramaniam et al., 2000; Hu et al., 2011). Moreover, the conserved outer membrane protein A was found to offer significant protection against homologous and heterologous virulent strains (Zhai et al., 2013).
Recently, the presence of genetically diverse R. anatipestifer in Kerala, India has been confirmed by biochemical and molecular methods (Priya et al., 2008; Pala et al., 2013; Pala and Radhakrishnan, 2014). The objective of the present study was to reveal the molecular characterization of outer membrane protein A gene (ompA) from different isolates of Kerala as this could explore the possibility of using ompA of R. anatipestifer for developing accurate and prompt control and diagnostic strategies.
Materials and Methods
Bacterial Isolates
The well characterized strains of R. anatipestifer maintained in the Department of Veterinary Biochemistry, College of Veterinary and Animal sciences, Mannuthy viz., KML-1, KML-2 and KML-3 were used for the study. Ready to use sterile sheep blood agar plates procured from Himedia Laboratories private limited, Mumbai were used for revival and subculture of the isolates. The bacterial isolates were identified based on morphology, cultural characteristics, growth on MacConkey’s agar, haemolysis on blood agar and biochemical tests like catalase and oxidase, indole production, gelatin liquefaction and ornithine decarboxylase activity.
Confirmation of R. anatipestifer Isolates by 16S rRNA Gene Based PCR
Final confirmation of the obtained isolates was done by amplifying partial region (665 bp) of 16S rRNA gene as per protocol described previously (Pala et al., 2013). Those isolates confirmed as R. anatipestifer were used for further analysis.
Amplification of ompA Gene
The supernatant collected after boiling and centrifugation of the colonies grown on blood agar was used as the source of DNA for PCR. The oligonucleotides OMP-F (5´ATGTTGATGACTGGACTTGGTCT3´) and OMP-R (5´CTTCACTACTGGAAGGTCAGACTT3´) (Yu et al., 2008) were used in a 25 μl reaction mixture containing 20 picomoles of each primer, 200 μmol l-1 each of dATP, dCTP, dGTP, and dTTP; 1.5 mmol l-1 MgCl2; and 1 U of Taq DNA polymerase (Genei, Bangalore). The thermal cycling profile consisted of denaturation at 94 °C for 1 min, annealing at 54 °C for 1 min, and extension at 72 °C for 1 min for 35 cycles followed by a final extension at 72 °C for 3 min. The PCR products were electrophoresed in 2% agarose for 1 h.
Cloning
The 1119 bp PCR product comprising the partial ompA encoding gene of all the three isolates were purified using GeneJETTM Gel Extraction Kit, (Fermentas Life Sciences, Lithuania) and cloned into InsT/Aclone vector (Fermentas Life Sciences, Lithuania). The recombinant clones obtained were selected in LB-Ampicillin agar plates containing IPTG and X-Gal and further screened by PCR for confirmation of the presence of the desired gene insert.
Sequence Analysis
Using the OMP-F and OMP-R primer set, the amplified products were sequenced at the DNA sequencing facility at Scigenom Pvt. Ltd., Cochin, Kerala using an automated DNA sequencer (Applied Biosystems, USA). The sequences were aligned and compared with the sequence data retrieved from GenBank. Sequence similarity search was performed using Basic Local Alignment Search Tool (BLAST) network provided by the National Centre for Biotechnology Information (NCBI). The sequences were subjected to multiple sequence analysis with the sequence data retrieved from GenBank. The sequences were translated to their respective aminoacid sequences using GeneTool Lite software and were subjected to phylogenetic analysis using the software MEGA version 6.0
Results
Bacterial Isolates
Freeze dried isolates of R. anatipestifer maintained in the Department of Veterinary Biochemistry, College of Veterinary and Animal Sciences, Mannuthy, viz., KML-1, KML-2 and KML-3 which were used for this study were revived by initial culture onto sheep blood agar plates. The mucoid, convex, greyish-white and non-haemolytic colonies were selected. All the isolates were non-motile, Gram-negative, coccobacillary, catalase and oxidase positive. None of the three isolates grew on MacConkey’s agar. All the isolates that were indole negative, ornithine decarboxylase negative and gelatin liquefaction positive were selected for further analysis by PCR.
Confirmation of R. anatipestifer Isolates by 16S rRNA Gene Based PCR
All the three isolates selected were successfully amplified with a 665 bp DNA product (data not shown) at an annealing temperature of 54°C confirming their identity as R. anatipestifer.
Amplification of ompA Gene
All the three isolates, on amplification using OMP-F and OMP-R primer pair, gave 1119 bp DNA products when observed on 2% agarose gel (Figure 1).
Cloning
The gel purified PCR products were cloned in the multiple cloning site of pTZ57R/T vector system. Following transformation, the recombinant E. coli appeared as white colonies on LB-Ampicillin agar plates. The presence of insert was again confirmed by colony PCR in all the three isolates.
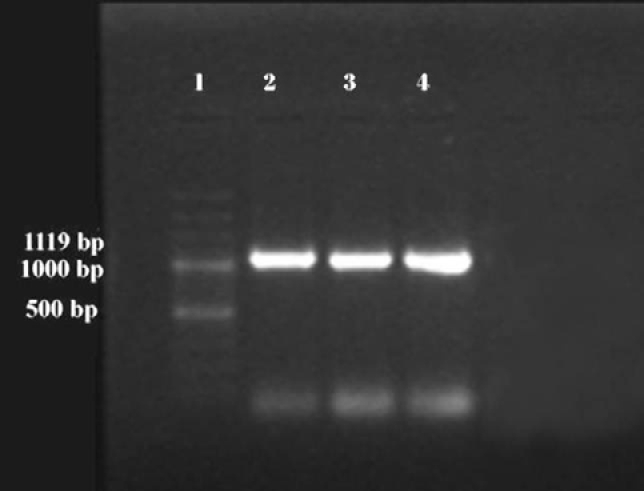
Figure 1: PCR amplified fragments of ompA gene of R. anatipestifer isolates: Lane 1) 100bp ladder; Lane 2) KML-1; Lane 3) KML-2; Lane 4) KML-3
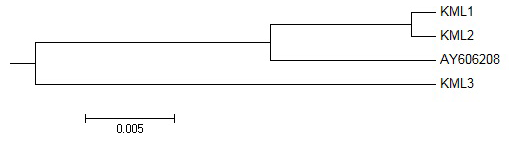
Figure 3: Dendrogram of ompA gene sequences of different isolates of R. anatipestifer depicting the relationship between each isolate and that of database sequence AY606208
Sequence Analysis
Sequencing of all the three isolates resulted 1119bp long nucleotide sequences in all the three isolates. On sequence similarity analysis using NCBI BLAST tool, all the sequences were found to be more identical to a Taiwanese strain of R. anatipestifer (Accession # AY606208). All the sequences have been submitted to GenBank and the allotted accession numbers are KX984363 (KML-1), KX984364(KML-2) and KX984365 (KML-3), respectively.
Multiple sequence alignment of the three isolates with the database sequence (AY606208) was performed using MEGA version 6.0 software (Figure 2, placed at the end). KML-1 showed 98.5% similarity to the databank sequence whereas KML-2 showed 98.1% similarity. KML-3 was the most variable among the three isolates showing 95.1% identity to AY606208. Comparison in between the isolates showed 96-99% identity. KML-1 and KML-2 were more similar, depicting 99.7% identity whereas KML-3 was 96.6 and 96.8 % similar to KML-1 and KML-2, respectively.
On phylogenetic analysis of the sequences using MEGA version 6.0 software, KML-1 and KML-2, which were more identical, were found to be clustered together on sub braches of one main branch while KML-3 delineated itself on a separate branch (Figure 3).
The nucleotide sequences were translated to respective aminoacid sequences using GeneTool Lite software. On analysis of the derived aminoacid sequences, it could be observed that the three isolates showed more variation towards the N-terminal compared to that of the C-terminal. The aminoacid sequences of KML-3 were found to be more different than the other two (Figure 4).

Figure 4: Predicted aminoacid sequences of different R. anatipestifer isolates (regions with sequence variations are shown in red colour)
Discussion
Riemerella anatipestifer is important in veterinary medicine as the causative of epizootic New Duck disease. So far at least 21 serotypes have been identified. The occurrence of more than one R. anatipestifer serotype in infected ducks at any one time and changes in serotypes from year to year within a single farm have been reported. There are strong variations of virulence among different serotypes of R. anatipestifer, and even within a given serotype. Vaccines based on inactivated bacteria confer some protection against homologous strains or serotypes, but bacterins prepared from heterologous serotypes do not provide cross-protection. Little progress has been made towards developing a subunit vaccine against R. anatipestifer and only the outer membrane protein gene, ompA, has been characterized to date. Outer membrane proteins play an important role in virulence and induce strong antibody responses that are bactericidal, opsonic or protective. Outer membrane proteins are therefore suitable candidate proteins for vaccine development as well as for the development of specific and sensitive diagnostic tools. Among the outer membrane proteins, Outer membrane protein A (OmpA) plays important role in maintenance of the structural integrity of the cell envelope, in bacterial conjugation, in adhesion/invasion and serum resistance etc. Keeping in mind the importance of utilizing ompA in developing control and diagnostic strategies to curb the menace posed by R. anatipestifer to the ducks of Kerala, the study was designed to characterize the ompA gene of R. anatipestifer isolated from ducks of Kerala.
The sequence data obtained from sequencing of the cloned PCR products provided the partial structural organization of the transcription unit. In all the isolates, amplicons of size of about 1100 bp were observed in 1.5 % agarose gel using 100 bp plus molecular size marker as a standard. Yu et al. (2008) amplified a product of the same size from the ompA of R. anatipestifer gene which encodes 42 kDa outer membrane protein (ompA) using the same primer. The amplicons were cloned in T/A vector and sequenced and the interpretation of results provided about 1100 bp long sequences. The obtained sequences were analysed using BLAST tool of NCBI which depicted that all sequences were about 95-99% similar to the database sequence AY606208 which belongs to a Taiwanese strain of R. anatipestifer.
The comparison of sequence of KML-1, KML-2 and KML-3 with the database sequence showed that most of sequence variations were seen in the region of 45 to 200 bp unlike the previous reports where the variations were mainly concentrated around 600-830bp region (Tsai et al., 2005; Yu et al., 2008). The aminoacid sequences derived from the nucleotide sequences were blasted using NCBI BLAST tool which showed 91-97% similarity to ompA protein of R. anatipestifer. Majority of the variations were found to be between 16-50 aminoacid residues which contribute to the formation of the outer membrane protein β barrel domain.
Analysis of nucleotide sequence of ompA gene of R. anatipestifer revealed that it encodes a protein of 387 amino acids with a molecular mass of 42 kDa (Subramaniam et al., 2000). The C-terminal half contains the characteristic OmpA-like domain, a stretch of 45 amino acids which shows high homology to outer membrane proteins of many gram-negative bacteria. The rest of the protein, especially the N-terminal amino acid sequence, shows no similarity to other outer membrane proteins. This is characteristic of OmpA proteins. As the sequence analysis of the gene showed no much variation among the isolates, the ompA gene based PCR could be utilised as a diagnostic tool to confirm the presence of R. anatipestifer from clinical samples.
The ompA gene based PCR could be utilised as a diagnostic tool to confirm the presence of R. anatipestifer from clinical samples since there is no significant variation in the make-up of the gene in between various isolates of the organism. Though the organism has been reported across the world in different species of poultry and the economic losses imparted on the farmers are huge, a complete picture of the virulence factors of the organism is not yet available and a little is known about its pathogenesis. A whole genome analysis of the organism which has been recently reported could accelerate the explorations in this regard (Zhou et al., 2011). A comprehensive knowledge on the genetic makeup of the organism could assist in adopting prompt diagnostic and control strategies.
Acknowledgments
The work was supported by the research grant from the Department of Animal Husbandry, Government of Kerala, India. The authors are grateful to Dr. Mahesh Mahendran, Veterinary Surgeon, Avian Disease Diagnostic Laboratory, Thiruvalla, Kerala, India for expert technical guidance.
Conflict of Interest
The authors have no conflict of interest.
authors’ contribution
Ciby Somu standardized and performed the molecular studies while Shonima Pala isolated and characterized the microorganism. Uma Radhakrishnan designed the study, analysed the results and prepared the manuscript.
References






