Advances in Animal and Veterinary Sciences
Research Article
Molecular Detection and Phylogenetic Analyses to Evaluate the Evolutionary Pattern of N, P, and G Gene Sequences of Some Recent Rabies Virus (RABV) Isolates from Elwadi- Elgedid Province in Egypt.
Michael Salheen1, Fouad S. El-mayet2*, Zeinab R. Aboezz2, Ayman S. El-Habbaa2, Marcus I. Yanni1, Saad S.A. Sharawi2
1Virology Department, Animal Health Research Institute (AHRI), ARC, Giza, Egypt; 2Virology Department, Faculty of Veterinary Medicine, Benha University, 13736, Kaliobyia, Egypt.
Abstract | Rabies is a fatal zoonotic infection caused by negative-strand RNA-virus classified in the genus Lyssavirus, family Rhabdoviridae of the order Mononegavirales. Despite the importance of the RABV for human and animal health, little is known about the spread of the virus in different animal populations in Egypt. Therefore, the current study aimed to identify and analyze the nucleotide sequences of nucleoprotein (N), phosphoprotein (P) and Glycoprotein (G) genes of the rabies virus from 10 suspected rabid cases of cattle and foxes from Elwadi- Elgedid province, which collected in 2017-2019. Specimens from the brains of suspected animals with rabies were collected and identified using direct FAT and RT-PCR. Three positive samples were further subjected to sequencing the partially amplified N, P, and G genes. The phylogenetic analysis was performed concerning the Egyptian rabies virus isolates, vaccinal strains and other sequences from the neighbouring countries available on the GenBank. It was found that the nucleotide identity in comparison to our new Egyptian isolates of Elwadi- Elgedid decreases by time at the level of three genes of previously isolated strains. Our results revealed that these new RABV isolates were more homologous to each other and were genetically related to Egyptian dog rabies, which previously identified two decades ago. The studied vaccinal strains were of low nucleotide identity with the new isolates. They also share some similarities with the RABV isolates in the neighbouring endemic regions in the Middle East. Furthermore, these data extend our knowledge of the relatedness and genetic variation of RABVs circulating in Egypt.
Keywords | Molecular analysis, nucleoprotein (N), genes, Rabies, Elwadi- Elgedid.
Received | June 30, 2021; Accepted | July 01, 2021; Published | October 01, 2021
*Correspondence | Fouad S El-Mayet, Virology Department, Faculty of Veterinary Medicine, Benha University, 13736, Kaliobyia, Egypt; Email: fouad.saad@fvtm.bu.edu.eg
Citation | Salheen M, El-Mayet FS, Aboezz ZR, El-Habbaa AS, Yanni MI, Sharawi SSA (2021). Molecular detection and phylogenetic analyses to evaluate the evolutionary pattern of N, P, and G gene sequences of some recent rabies virus (RABV) isolates from Elwadi- Elgedid province in Egypt. Adv. Anim. Vet. Sci. 9(11): 1989-1994.
DOI | http://dx.doi.org/10.17582/journal.aavs/2021/9.11.1989.1994
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2021 El-Mayet et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Rabies is a world-widely distributing zoonotic disease with highly acute fatal encephalomyelitis in humans. The etiological agent is the rabies virus (RABV) which belongs to the genus Lyssavirus in the family Rhabdoviridae. It has a linear non-segmented single-stranded RNA of a negative sense of 12 kb length (MacLachlan et al., 2016). The genome has 58-nt of 3′ leader and 70-nt 5′ trailer. the genome composed of five genes Nucleoprotein (N) gene (nt 59 to 1483), phosphoprotein (P) gene (nt 1486 to 2475), matrix protein (M) gene (nt 2481 to 3283), glycoprotein (G) gene (nt 3289 to 5355), L gene (nt 5380 to 11853). Four variable-length intergenic regions separate these five genes; N-P, P-M, M-G, and G-L contain 2, 5, 5, and 24 nt, respectively (Wunner et al., 1988, Luo et al., 2012). Like other RNA viruses, there is a higher average of mutations in lyssaviruses due to a lack of proofreading activity of the polymerase (L) (Steinhauer and Holland, 1987). There are seven genotypes of lyssavirus defined; rabies virus (genotype 1) is responsible for classical rabies in terrestrial mammals and distributed worldwide except Antarctica and some islands, Lagos bat virus (genotype 2), Mokola (genotype 3), Duvenhage virus (genotype 4), European bat lyssavirus type 1 (genotype5), European bat lyssavirus type 2 (genotype 6) and Australia bat lyssavirus (genotype 7) (Childs and Real, 2007).
The viral glycoprotein (G) is contributed to RABV pathogenicity. It has a significant role in viral neuroinvasiveness. Most of the molecular studies of the rabies virus were based on the glycoprotein (G) gene as a lower rate of nucleotide substitution gets occurred (Faber et al., 2004; Troupin et al., 2016). Also, the N gene sequence is the highly conserved sequence among the other viral genes so, these sequences used in genetic typing and evolutionary studies. Moreover, their a relative lower variation among reservoir-associated variants and geographic lineages (Bourhy et al., 1993; Kissi et al., 1995). In addition to; the multifunction of phosphoprotein (P), it antagonizes the type I interferon (IFN)-mediated antiviral responses helping in evasion of host innate immunity and plays an essential role in viral RNA synthesis (Wunner and Conzelmann, 2013; Blondel et al., 2002).
Several previous molecular and phylogenetic analyses were applied on individual genes, but no studies on multiple viral genes were done in Egypt decades ago. Therefore, the purpose of the present study was to provide insight into the genetic variability, and phylogenetic relatedness of the rabies virus isolates derived from rabid cattle and foxes in Egypt compared with the other strains of RABV vaccinal strains available on GenBank.
Materials and methods
Collection and processing of samples
Ten brain samples were collected from suspected rabid animals (cattle and foxes) in Elwadi- Elgedid province (Figure 1). Each brain sample was processed as soon as it was received for laboratory diagnosis of the rabies virus. Impression smears were made from the hippocampus for direct fluorescent antibody techniques (FAT). The supernatants of brain samples homogenate were used to inoculate BHK-21 cells (Chhabra et al., 2007).
Direct fluorescent antibody technique (dFAT)
It was carried out as described previously by (Dean et al., 1996). The Brain impression smears, and inoculated BHK-21 cells were air-dried and fixed in chilled acetone, then stained with FITC- conjugated anti-RABV monoclonal antibodies (Catalog # MA1-90618, Invitrogen). The slides were kept in a humidified chamber for 45 minutes at 37 ◦C. and washed thoroughly with PBS for 15 min three times, then mounted with buffered glycerin and covered with a coverslip for examination by fluorescent microscope (Olympus, Japan).
Nucleic acid extraction and RT-PCR assay
Ten grams of positively infected brain samples were used for RNA extraction using the QIAamp viral RNA mini kit (Cat. No: 52904, Qiagen, Germany). RT-PCR assay was performed on the extracted RNA using Qiagen one-step RT-PCR kit with three pairs of specific primer sets were used as in Table (1). These oligonucleotides primers were supplied by (Metabion, Germany). The PCR products were analyzed by electrophoresis on 1.5% agarose gel.
Sequencing of the amplified products and analysis of sequencing data.
Gel purified PCR products for N, P and G genes were sequenced using the ABI PRISM BigDye terminator cycle V3.1 sequencing kit (Life Technologies, USA). The obtained sequence data were analyzed in MEGA X using ClustalW (Kumar et al., 2018).
Gene Bank accession numbers
The obtained sequences for EgyWN17, EgyWN18 and EgyWN19 RABV isolates were assigned accession numbers MT036064, MT036065, MT036066, respectively for the N gene. While the P gene sequences were given accession numbers MT036067, MT036068, MT036069 and The G gene sequences were allocated accession numbers MT036061, MT036062, MT036063, respectively.
Table 1: Primers sequences, target genes, amplicon sizes.
| Target gene | Primer sequence (5'-3') | Target gene | Size of amplicons (bp) | Reference |
| P | 5´-ttggtacscatcccaagtatga -3´ | P | 882 |
|
| 5´-tcttgcatgattttgcttagcttgtc -3´ | ||||
| N | 5´-atggatgccgacaagattgt -3´ | N | 684 | |
| 5´-cccactctgattgccgaata -3´ | ||||
| G | 5´-agtagagggaagagagcatcca -3´ | G | 590 | |
| 5´-gaggataggaacaactccat -3´ |
Results
Identification of the rabies virus
The rabies antigen in the infected brain tissue was detected by performing dFAT on both infected brain tissue and inoculated baby hamster kidney (BHK-21) cells. The obtained results approved the presence of RABV in the infected cells and brain specimens from the diseased cows and foxes, while the normal brain and uninfected cells showed a negative reaction (Figure 2).
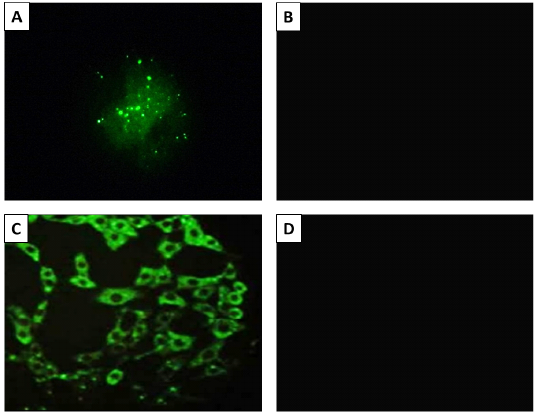
Figure 2: The result of fluorescent antibody technique (FAT) Panel A; Positive (bright green apple fluorescence) on brain impression smear. Panel B; Negative control sample, Panel C; Positive fluorescence on BHK-21 cells inoculated with Rabies virus. Panel B; Negative control (uninfected cells). 400X magnification.
Molecular identification of viral samples by reverse transcriptase RT-PCR
The PCR products obtained from the amplification process of the extracted RNA were as follow for nucleoprotein (N) gene with expected correct size (684 bp), Phosphoprotein(P) gene with (882 bp) and Glycoprotein (G) gene with (590 bp) when compared with each positive control samples (Figure 3).
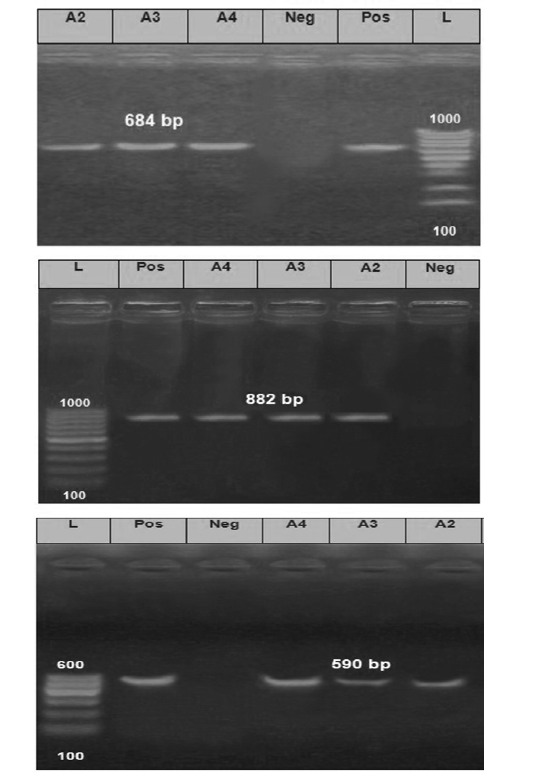
Figure 3: The Electrophoretic pattern of the RT- PCR products to the brain samples (Lab codes; A2, A3 and A4) on the gel electrophoresis (1.5%), Panel A; G gene (590 bp), Panel B; N gene (684bp) and Panel C; P gene (882 bp). Positive and negative controls are included.
Nucleotide sequencing and phylogenetic analysis of sequencing data
Our sequence data were compared with 30 different Egyptian rabies virus isolates, Egyptian vaccinal strains, and other nearby geographic location isolates for their N, P and G gene sequences. The phylogenetic analysis demonstrated that EgyWN17 (MT036064) and EgyWN18 (MT036065) were completely identical to each other with 99.7% of homology with EgyWN19 (MT036066) at the N gene sequence level. Moreover, these sequences were very close to rabies isolates in 2013 from the same locality (Elwadi- Elgedid). And with more than 98% of identity with horse RABVs with accession no. (KP119663 & KP119662) those were isolated from Cairo. While as; multiple nucleotide sequence alignment with the neighbouring countries rabies isolates revealed 92% of identity with RABVs of Israel and Turkey (accession no. DQ837455, DQ837393, DQ837480 & DQ837475), 90 % of identity with Sudan RABVs and <90 % with Saudi Arabia, Algeria and Italy viruses (Figure 4).
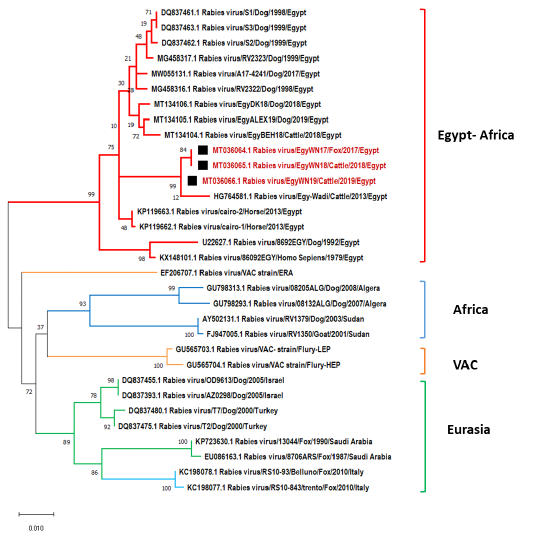
Figure 4: Phylogenetic tree using maximum likelihood method based on nucleoprotein (N) gene nucleotide sequences of our new RABV isolates from Elwadi- Elgedid (indicated by black squares) with other sequences of rabies viruses retrieved from the GenBank database. Numbers at the internal nodes represent the bootstrap probabilities (1000 replicates). The scale bar (0.01) means nucleotide changes or substitutions per site.
At the level of the P gene, EgyWN17 (MT036067), EgyWN18 (MT036068) and EgyWN19 (MT036069) were approximately identical to each other as 98.9% to 99.6% of similarity. They were significantly closer by 96.5% of homology with the old Egyptian isolates, and recent Egyptian isolates from Behera, Dakahlia and Alexandria While, the nucleotide identity with Egy79 (KX148101) was 95%. By comparing our P gene sequence data with previously non-Egyptian isolates, it was found that the Israeli strain (KF154998) was the highest (>95%) than the other Asian, African and European Isolates (<90 %) (Figure 5).
The obtained G gene sequences (MT036061, MT036062 and MT036063) were completely identical to each other. They had a higher degree of identity range from97-97.7%
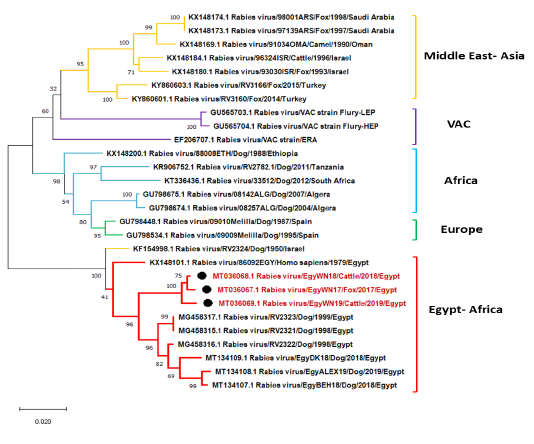
Figure 5: Phylogenetic tree using maximum likelihood method based on phosphoprotein (P) gene nucleotide sequences of our new RABV isolates from Elwadi- Elgedid (indicated by black circles) with other sequences of rabies viruses retrieved from the GenBank database. Numbers at the internal nodes represent the bootstrap probabilities (1000 replicates). The scale bar (0.02) means nucleotide changes or substitutions per site.
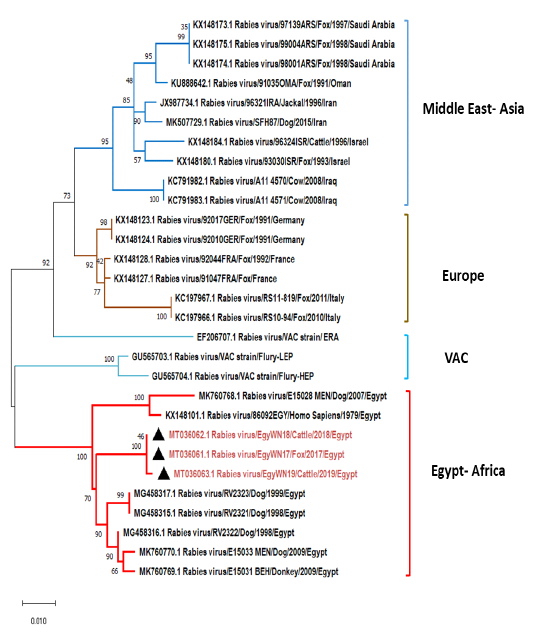
Figure 6: Phylogenetic tree using maximum likelihood method based on glycoprotein (G) gene nucleotide sequences of our new RABV isolates from Elwadi- Elgedid (indicated by black triangles) with other sequences of rabies viruses retrieved from the GenBank database. Numbers at the internal nodes represent the bootstrap probabilities (1000 replicates). The scale bar (0.01) means nucleotide changes or substitutions per site.
to the following Egyptian isolates (accession no. MG458315, MG458316, MG458317, MK760770, and MK760769). While MK760769 and KX148101 have shared 94.9% and 96% nucleotide identity, respectively (Figure 6). On comparing with some previously non-Egyptian isolates of nearby geographic locations, it was found that the divergence percentage was 6-9% based on pairwise distance analysis.
On studying the pattern of pairwise distances for ERA vaccinal strain, Flurry LEP strain, and Flurry HEP strain at the level N, P, and G genes to our new Egyptian isolates from Elwadi- Elgedid, it was ranged from 8.7-9.5% for N gene and from 12.4 – 12.8% for P gene, while it had a narrow range from 4.2-5.3% for G gene (Table 2).
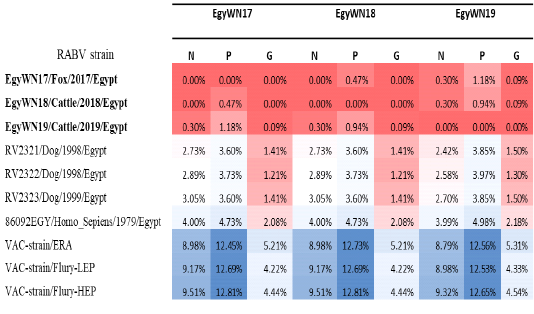
Table 2: Pairwise distance percentages of the nucleotide sequences of N, P and G genes of the three new Egyptian isolates in relation to themselves and other Egyptian and vaccinal strains retrieved from the GenBank database. The highest related genes have a deep red color that converts to light red, light blue and deep blue with increasing distance.
Discussion
In Egypt, rabies is an enzootic viral disease-causing public severe health concerns. The persistence of RABV in various animals might be attributed to the significant population of roaming stray dogs. Moreover, a lack of effective control measures. Rabies viral infection needs immediate investigation for proper decisions as slaughter or immunization. dFAT and RT- RCR were used in the following study as they are rapid and efficient methods. They were used for laboratory investigation of ten brain samples of suspected cattle and foxes collected from Elwadi- Elgedid province in Egypt (El-Tholoth et al., 2015; Zhang et al., 2011). There is no need for virus propagation on cell culture as RT-PCR and dFAT can detect RABV in fresh brain tissue of infected animals. This might influence inappropriate measures for control (Singh et al., 2017).
The phylogenetic analysis of the new viral isolates (EgyWN17, EgyWN18 and EgyWN19) revealed a high degree of identity with the former Egyptian isolates that originated from different species. This was reasonable as the previous finding indicated that RABV isolates from different species clustered according to their geography and not their species (David et al., 2007; Nagarajan et al., 2006; Reddy et al., 2011). The partial nucleotide sequences of our new isolates for nucleoprotein (N) (684bp), phosphoprotein (P) (882bp) and glycoprotein (G) (590bp) genes; demonstrated an extremely close relation with previously isolated RABV from the same geographical region. This might be attributed to their originating from a common predecessor spread among different provinces that were agreeable with (Bourhy et al., 1993; Kissi et al., 1995).
Otherwise, our viral isolate sequences revealed distinctive identity with RABV from neighbouring countries as Israel, Turkey, and Sudan. This indicates the possibility of RABV transmission from and to our neighbouring countries (Body et al., 2014; Nagarajan et al., 2009). Regarding the Egyptian RABV vaccinal strains, they were far from our new isolates based on the similarity ratio in N, P, and G gene nucleotide sequence. Despite this, the P and N genes considered the most conserved among the other virus genes, and relatively lower variation among reservoir-associated variants and geographic lineages (Bourhy et al., 1993; Kissi et al., 1995).
From this study, we could conclude that the brain specimens of cattle and foxes revealed the presence of new RABV isolates that are relatively similar to RABVs from some neighbouring Middle East countries as the unrestricted movement of foxes and stray dogs possibly help in the transmission of rabies virus from and to nearby geographic regions.
Acknowledgements
The authors would like to thank the Virology laboratory unit in the Animal Health Research Institute (AHRI) for providing the facilities for conducting the study. The authors did not receive any funds for this study.
Conflicts of Interest
The authors declare no conflict of interest.
Author Contributions
The conceptualization of this study was performed by S. Sharawi, M. Yanni., and M. Salheen. Investigation and Methodology were performed by M. Salheen, S. Sharawi and M. Yanni. Data curation and phylogenetic analyses were performed by F. El-mayet, A. El-Habbaa and Z. Aboezz. The first draft of the manuscript was written by A. El-Habbaa, M. Yanni and M. Salheen. Reviewing and F. El-mayet, and M. Salheen performed editing of the manuscript. All authors have read and agreed to the published version of the manuscript.
References