Advances in Animal and Veterinary Sciences
Research Article
Analysis of the Genetic Structure of the Hereford Population Bred in Kazakhstan
Askar Myrzahmetovich Nametov1*, Indira Saltanovna Beishova2, Alexandr Mikhailovich Kovalchuk2, Tatyana Vladimirovna Poddudinskaya2, Alena Valentinovna Belaya3
1Non-Commercial Joint-Stock Company, Zhangir Khan West Kazakhstan Agrarian Technical University; 2Republican State Enterprise on Right of Economic Conducting, Kostanai State University A. Baitursynov, Ministry of Education and Science of the Republic of Kazakhstan; 3Educational Establishment, Belarusian State Pedagogical University Maxim Tank, Belarus.
Abstract | Animal breeding is the main agricultural branch in the Republic of Kazakhstan, which is facing important tasks like providing food for the population and raw materials for the consumer industry. For this purpose, not only the conditions of animal feeding and the technology of fodder preparation are to be improved, but also new promising methods of increasing animals productivity are to be developed and introduced, and brought into conformity with the requirements that involve new types of animals, the physiology of which is determined by higher productivity and adaptability to the intense conditions of the environment and exploitation. Today’s DNA technologies allow speeding up the rate of breeding and reducing the financial costs in course of classic animal breeding. MAS-breeding relies on the information about phenotypic manifestations of alleles of the genes that are responsible for quantitative traits (candidate genes) and allows assessing the genetic potential of animal productivity in the early stages of postnatal development. The genes in the somatotropin cascade are polymorphic; a wide range of alleles has been identified in cattle, which are of interest for MAS breeding as genetic markers for economically useful traits. Since Hereford cattle is currently being massively purchased within the limits of large-scale government programs, it is important to study the association of the alleles of the genes in the somatotropin cascade with the traits of meat productivity in the cattle of this breed.
Keywords | Polymorphism, Meat productivity, Hereford, Breeding, Genes of the somatotropin cascade
Received | June 12, 2019; Accepted | August 30, 2019; Published | October 15, 2019
*Correspondence | Askar Myrzahmetovich Nametov, Non-Commercial Joint-Stock Company, Zhangir Khan West Kazakhstan Agrarian Technical University; Email: nametov.a.m@mail.ru
Citation | Nametov AM, Beishova IS, Kovalchuk AM, Poddudinskaya TV, Belaya AV (2019). Analysis of the genetic structure of the hereford population bred in Kazakhstan. Adv. Anim. Vet. Sci. 7(s1): 71-77.
DOI | http://dx.doi.org/10.17582/journal.aavs/2019/7.s1.71.77
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2019 Nametov et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
Today, over 22 thousand animals of the Hereford breed are present in 13 regions of Kazakhstan (Hereford Breed, n.d.). The Hereford breed is very widespread, it is bred in the United States, Canada, Australia, Europe, and the CIS countries. Herefords have valuable qualities: fast economic and physiological maturity, good reproductive ability. Hereford meat has good taste and cooking qualities: it is tender, juicy, high-caloric due to its natural ability to hold water and to the presence of intramuscular fat.
In the Republic of Kazakhstan, Hereford cattle are being intensively reproduced, therefore, studying the somatotropin cascade alleles associations with traits of meat productivity is an urgent task for the agricultural sector.
Assessment of animals by genetic markers becomes more efficient if it includes genes of the same physiological path because in this case the expression of one gene affects the expression of all other genes. Therefore, in analyzing the combined effect of polymorphisms on the analyzed traits, paired combinations with a potentiating effect are found (Belaya and Batin, 2012; Zhang et al., 2018; Ghoreishifar et al., 2019).
Of great interest for improving the meat productivity of cattle are the genes of the somatotropin cascade, the protein products of which are key links in the humoral chain involved in the process of lactation and in the growth and development of mammals (bGH, bGHR, bIGF-1) (Hines, 1990; Blackburn et al,. 2017). Therefore, studying polymorphisms of these genes is promising from the point of view of searching for the markers associated with the traits of both dairy and meat productivity of cattle.
It is known that the growth hormone and a number of other proteins (directly or indirectly required for its functioning) provide various molecular and cellular effects that ultimately result in organism development and growth. These proteins constitute a kind of axis or a system that launches and monitors the set of metabolic processes resulting in growth and associated with cellular differentiation.
Resolving theoretical and applied problems of farm animals genetics is impossible without the use of highly polymorphic reliable molecular-genetic marker systems that allow assessing intra- and interspecific genetic variability of the animals, the peculiarities of the microevolutionary processes that occur under the influence of breeding work, and identifying the most informative loci on the genome that determine high productivity and resistance of the animals to the disease (Beishova and Poddudinskaya, 2017; Chen et al., 2018a, 2018b).
MATERIALS AND METHODS
Molecular genetic studies and analysis of the obtained results were performed on the basis of the Department of the Molecular Genetic Research of the Testing Laboratory for Foodstuffs Production at the Research and Innovation Center of KSU n.a. A. Baitursynov.
The research study was made on 200 Hereford animals bred in Kazakhstan (Limited Liability Company Arystan RK).
Whole blood was the material for the study. DNA was isolated from the blood using the commercial Pure Link Genomic DNA Kits (manufactured by Thermo Fisher Scientific, USA) according to the manufacturer’s instructions.
The composition of the reaction mixture for the studied polymorphic loci of somatotropin cascade genes was the following:
The primer sequences and the PCR conditions for analyzing each polymorphism are shown in Table 1.
PCR products were detected in 2 % agarose gel for one hour and in 3 % agarose gel for two hours at 110V.
Polymorphism of the nucleotide sequence of the bGH gene in exon 5 was analyzed with the use of the AluI restrictase. Polymorphism is determined by transition C→G resulting in the replacement of the leucine amino acid with valine in a protein sequence. A recognition site for the AluI restrictase is the AD↓ST sequence. The allele recognized by the enzyme contains nucleotide C, and is marked as bGH-AluIL. In case of the presence of the G nucleotide, the restriction site disappears; such an allele is marked as bGH-AluIV. The resulting electrophoresis pattern is shown in Figure 1.
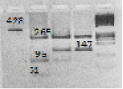
Figure 1: Electrophoretogram of DNA-typing of the bGH-AluI polymorphism (Skinkytė et al., 2005); Track 1 – amplificate 428 of the nucleotide pairs in a fragment of the bGH-AluI gene; track 2 – fragments of restriction 265, 96, 51 p.n. (16. p.n. are not visualized) corresponding to the bGH-AluILL genotype; track 3–fragments of restriction 265, 147, 96, 51 p.n. corresponding to the bGH-AluILV genotype; track 4–fragments of restriction 265 and 147 p.n. corresponding to the bGH-AluIVV genotype; and track 5– molecular masses marker O’Range RulerTM DNA Ladder, Fermentas, Lithuania. Electrophoresis was performed in 2% agarose gel (SeaKem LE Agarose, Lonza, USA).
Table 1: Individual characteristics of the PCR conditions for the studied polymorphic loci of somatotropin cascade genes.
| Polymorphism | Amplification conditions | Primer sequences |
|
bGH-AluI |
95°C – 10 min; (94°C – 30 sec; 61°C – 50 sec; 72°C – 30 sec) x 40 cycles; 72°C – 10 min |
F: 5′-ccgtgtctatgagaagc-3′ |
|
R: 5'-gttcttgagcagcgcgt-3′ |
||
|
bGHR-SspI |
95°C – 3 min; (95°C – 30 sec; 61°C – 30 sec; 72°C – 30 sec) x 30 cycles; 72°C – 10 min; 12°C – 5 min |
F: 5′- aatacttgggctagcagtgacaatat-3′ |
|
R: 5′-acgtttcactgggttgatga-3′ |
||
|
bIGF-1-SnaBI |
95°C – 5 min; (95°C – 30 sec; 62°C – 30 sec; 72°C – 30 sec) x 40 cycles; 72°C – 10 min |
F:5′-attacaaagctgcctgcccc-3′ |
|
R: 5′- accttacccgtatgaaaggaatatacgt-3′ |
The length of the amplified fragment of the bGH gene is 428 p.n. The length of the fragments after the restriction is 265, 147, 96, 51, and 16 p.n. The electrophoretogram shows the variants of bands of a certain length that are characteristic for genotypes of 265, 96, 51 p.n. (16 p.n. are not visualized) corresponding to the bGH-AluILL genotype; of 265 and 147 p.n. corresponding to the bGH-AluIVV genotype; and three bands of 208, 172, and 35 p.n. (the bGH-AluILV genotype). Fragment of the 35 BP restriction is not visualized on an agarose gel.

Figure 2: An electrophoretogram of DNA typing of the bGHR-SspI polymorphism (Zhang 1993); Track 1 – PCR product of the 182 p.n. fragment of the bGHR-SspI gene; tracks 2, 3, and 4 – a fragment of the 158 p.n. restriction corresponding to the bGHR-SspIFF genotype; track 5 – a fragment of the 182 p.n. restriction corresponding to the bGHR-SspIYY genotype; and track 6 – fragments of the 182 and 158 p.n. restriction corresponding to the bGHR-SspIFY genotype. The fragment of 24 p.n. is not visualized. The authors used molecular mass marker O’Range Ruler TM 50 bp DNA Ladder, Fermentas, Lithuania. The positions of specific bands on the gel are shown by arrows. Electrophoresis was performed in 2% agarose gel (SeaKem LE Agarose, Lonza, USA).
Polymorphism of the nucleotide sequence of the bGHbGHR gene in exon 8 was analyzed with the use of the SspI restrictase. The SspI restrictase recognizes the T→A transition in exon 8. This SNP causes a substitution of the polar, although uncharged, tyrosine residue instead of neutral phenylalanine at protein position 279. The restrictase recognition site is the AAT↓ATT sequence. The amplificate dissected with the enzyme contains nucleotide T corresponding to the bGHR-SspIF allele. In the presence of A-nucleotide, the restriction site disappears; such an allele is marked as bGHR-SspIY. The length of the amplified fragment of the bGHR gene is 182 p.n. The length of the fragments after the restriction is 158 and 24 p.n. The electrophoretogram shows variants of bands of a certain length that are characteristic for genotypes: one band of 182 p.n. (the bGHR-SspIYY genotype), two bands of 158 and 24 p.n. (the bGHR-SspIFF genotype); and three bands of 182, 158, and 24 p.n. (the bGHR-SspIFY genotype). The fragment of 24 p.n. is not visualized on agarose gel (Figure 2).
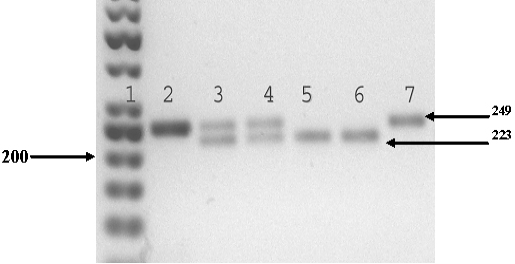
Figure 3: Electrophoretogram of DNA typing of the bIGF-1-SnaBI polymorphism (Kenny et al., 2011); Track 1 – molecular mass marker O’Range Ruler TM 50 bp DNA Ladder, Fermentas, Lithuania; track 2 – PCR product of the 249 p.n. fragment of the bIGF-1-SnaBI gene; tracks 3 and 4 – fragments of the 249 and 223 p.n. restriction corresponding to the bIGF-1-SnaBIAV genotype; tracks 5 and 6 – a fragment of the 223 p.n. restriction corresponding to the bIGF-1-SnaBIAA genotype; and track 7 – a fragment of the 249 p.n. restriction corresponding to the bIGF-1-SnaBIVV genotype. The fragment of 26 p.n. is not visualized. The positions of specific bands on the gel are shown by arrows. Electrophoresis was performed in 2 % agarose gel (SeaKem LE Agarose, Lonza, USA).
The polymorphism of the nucleotide sequence of the 1 bIGF-1 insulin-like growth factor gene in the area of the P1 promoter region is identified as T→S transversion. This substitution is recognized by the SnaBI restrictase. Two alleles have been recognized: bIGF-1-SnaBI allele (nucleotide T) dissected with the enzyme, and bIGF-1-SnaBI allele with nucleotide C characterized by the absence of the restriction site. A fragment of the bIGF-1 gene with a length of 249 p.n. is amplified. The length of the fragments after the restriction is 223 and 26 p.n. The electrophoretogram shows variants of bands of a certain length that are characteristic for genotypes: one band of 249 p.n. (the bIGF-1-SnaBIВВ genotype), two bands of 223 and 26 p.n. (the bIGF-1toSnaBIAA genotype); and three bands of 249, 223, and 26 p.n. (the bIGF-1-SnaBIAV genotype). The fragment of 26 p.n. is not visualized on agarose gel (Figure 3).
The research of the genetic structure of the analyzed Hereford populations involves a comparison of the samples by the distribution of allelic variants of the somatotropin cascade genes, as well as the assessment of genotypes distribution conformity to the theoretically expected one in accordance with the Hardy-Weinberg’s law.
Genotypes frequencies are determined by direct counting.
The relative occurrence rates of the alleles of the studied genes are determined by Formula 1:
where; N1 is the number of homozygotes in the studied allele, N2 is the number of heterozygotes, and n is the sample size (Mikhailova and Batin, 2012).
The statistical error of relative allele occurrence rates is calculated using Formula 2:
where Q is the relative occurrence rate of the studied allele, and n is the sample size (Rokitsky, 1961).
The samples by the occurrence rates of allelic variants of the studied genes are compared with the help of criterion χ2, Formula 3. The number of degrees of freedom = 1:
Where; Ho is the observed allele occurrence rates, and Ne is the expected alleles occurrence rates (Rokitsky, 1961).
If the expected values at least in one class are less than five, χ2 is calculated with Yates’s correction, Formula 4:
The correspondence of actual genotypes distribution to the theoretically expected one is in accordance with the Hardy-Weinberg’s law using criterion χ2, Formula 5. The number of degrees of freedom is one (the number of genotypes minus the number of alleles).
Where Ho is the observed frequency of genotypes, He is the expected occurrence rate of genotypes, AA = p2, AB = 2pq, and BB = q2 (Rokitsky, 1961).
If the expected values at least in one class are less than five, χ2 is calculated with Yates’s correction:
The allowed value for χ2 for one degree of freedom and 5 % significance level is 3.84 (Rokitsky, 1961).
RESULTS AND DISCUSSION
Analysis of the genetic structure of a population includes studying the nature of genotypes distribution, its correspondence to the theoretically expected one according to the Hardy-Weinberg’s law, as well as an analysis of the alleles occurrence rates distribution in the studied polymorphic somatotropin cascade genes.
The nature of the polymorphic gene bGH genotypes distribution is shown in Table 2.
Table 2: The polymorphic gene bGH genotype occurrence rates distribution in Hereford cattle (n = 198).
| Genotype | n observed | % of n | n expected | % of n |
χ2 |
|
bGH-AluIVV |
34 | 17.17 | 20 | 10.05 | 22.04 |
|
bGH-AluILV |
57 | 28.79 | 86 | 43.22 | |
|
bGH-AluILL |
107 | 54.04 | 93 | 46.73 |
Note: deviation of the observed genotype occurrence rates from the theoretically expected ones according to the Hardy-Weinberg’s law is veracious with χ2 ≥ 3.84.
Table 2 shows that in the studied sample, a statistically veracious deviation of genotypes occurrence rates distribution from the theoretically expected one according to the Hardy Weinberg’s law is observed. In particular, the excess of the number of both homozygotes is observed. The most common is bGH-AluILL genotype, its occurrence rate is 54.04 %. The 2nd place in terms of occurrence rate is taken by bGH-AluILV 28.79 %, and in the last place by bGH-AlulVV homozygotes, their occurrence rate is 17.17 %.
Comparing the inbreed data for the Herefords obtained by other authors studying populations of this breed in various countries, it should be noted that the deviation from Hardy-Weinberg’s law has been noted in most studies. However, its nature in all cases is different. For instance, the work of Krasnopiorova in studying the Lithuanian population of Herefords has shown 90 % occurrence rate of the bGH-AluILL genotype. At the same time, studies of T.A. Sedykh on the Russian population have shown the ratio of bGH-AluILL, bGH-AluILV and bGH-AluIVV genotypes of 57.7 %, 30.76 %, and 11.04 % respectively, which substantially correlates with the data of the authors.
The most likely explanation for the observed peculiarities of genotypes distribution is an association of the bGH-AluILL genotype with the economically useful traits in Hereford cattle, which, in course of artificial selection, results in a predominance of this genotype in the population.
The analysis of the distribution of the relative occurrence rates of alleles of polymorphic gene variants bGH, bGHR and bIGF-1 was performed based on the data shown in Table 3.
Table 3: The polymorphic gene bGHR genotype occurrence rates distribution in Hereford cattle (n = 200).
| Genotype | n observed | % of n | n expected | % of n |
χ2 |
|
bGHR-SspIYY |
24 | 12.00 | 29 | 14.43 | 1.83 |
|
bGHR-SspIFY |
103 | 51.50 | 94 | 46.77 | |
|
bGHR-SspIFF |
73 | 36.50 | 78 | 38.81 |
Note: deviation of the observed genotype occurrence rates from the theoretically expected ones according to the Hardy-Weinberg’s law is veracious with χ2 ≥ 3.84.
From Table 3 it follows that in the case of bGHR-SspI polymorphism, deviation from the expected distribution of alleles occurrence rates is not statistically veracious. The first place in terms of occurrence rates, in this case, is taken by bGHR-SspIFY heterozygotes, the second place by bGHR-SspIFF homozygotes, and the last place in the studied population by bGHR-SspIYY homozygotes. Their ratios amounted to 51.50 %, 36.50 %, and 12.00 %, respectively. Unfortunately, the data about the frequency rates of the genotypes of this polymorphism are absent for Herefords in the literature. However, it can be noted that some excess in the population of occurrence rates of bGHR-SspIFY genotypes is an evidence of positive association of the growth hormone receptor gene’s heterozygosity with the economically useful traits. It may also be assumed that homozygous bGHR-SspIFF genotype is preferred for some qualities of the phenotype, compared to bGHR-SspIYY genotype.
The nature of the polymorphic gene bIGF-1 genotypes distribution is shown in Table 4.
According to the data in Table 4, distribution of genotypes of this polymorphism in the population veraciously deviates from the theoretically expected one according to the Hardy-Weinberg’s law. In particular, there is an excess of the number of bIGF-1-SnaBIAV heterozygotes observed in the population (54.55 % of the observed, compared to the theoretically expected 47.74 % according to the Hardy-Weinberg’s law). This allows assuming the association of this genotype with some breeding advantage in animal carriers. Unfortunately, the literature contains no data about occurrence rates of the genotypes of this polymorphism in Herefords.
Table 4: The polymorphic gene bIGF-1 genotype occurrence rates distribution in Hereford cattle (n = 198)
| Genotype | n observed | % of n | n expected | % of n |
χ2 |
|
bIGF-1-SnaBIАА |
24 | 12.12 | 31 | 15.58 | 4.01 |
|
bIGF-1-SnaBIАВ |
108 | 54.55 | 95 | 47.74 | |
|
bIGF-1-SnaBIВВ |
66 | 33.33 | 73 | 36.68 |
Note: deviation of the observed genotype occurrence rates from the theoretically expected ones according to the Hardy-Weinberg’s law is veracious with χ2 ≥ 3.84.
The nature of the distribution of relative occurrence rates of AluI polymorphic alleles of the bGH gene in a population of Herefords bred in Kazakhstan is shown in the data in Table 5.
Table 5: Distribution of the relative occurrence rates of the AluI polymorphic alleles of gene bGH in a population of Herefords bred in Kazakhstan (Q ± SQ).
| Allele | Observed occurrence rates of alleles | Relative occurrence rates of alleles |
|
bGH-AluIL |
271 | 0.684 ± 0.002 |
|
bGH-AluIV |
125 | 0.316 ± 0.002 |
Table 5 shows that bGH-AluIL allele is the most widespread, compared to bGH-AluIV allele. These data coincide with the data obtained on the Russian population of Herefords studied by T.A. Sedykh. According to her data, the ratio of bGH-AluIL allele to bGH-AluIV allele is 0.684 to 0.316, which coincides with the data of the authors. According to N. Krasnopiorova, in studying the Lithuanian population of Herefords, the ratio of occurrence rates for bGH-AluIL and bGH-AluIV alleles is 0.9 to 0.1. This allows assuming that bGH-AluIV allele is associated with some undesirable characteristics, which in the conditions of the Lithuanian selection is expressed stronger than in the populations of Herefords bred in Russia and in Kazakhstan.
The results of assessing the distribution of relative occurrence rates for the SspI polymorphic alleles of bGHR gene in the population of Herefords bred in Kazakhstan are shown in Table 6.
The data in Table 6 show that the ratio of bGHR-SspIF alleles to bGHR-SspIY alleles corresponds to the distribution in terms of bGH-AluI polymorphism, while the genotype distribution, as discussed above, does not. In the earlier works, the authors assumed that an increased occurrence rate of heterozygotes in terms of polymorphism of the growth hormone receptor gene in comparison with the shares of heterozygotes in terms of polymorphism of the somatotropin gene was due to the fact that the presence of the two types of growth hormone receptor protein in the organism possibly involved additional mechanisms for mediating the physiological effect of somatotropin on target cells, and thus gave growth and meat productivity advantages to the animals that were heterozygous in terms of growth hormone receptor gene (Beishova et al., 2017).
Table 6: Distribution of the relative occurrence rates of the SspI polymorphic alleles of bGHR gene in the population of Herefords bred in Kazakhstan (Q ± SQ).
| Allele | Observed occurrence rates of alleles | Relative occurrence rates of alleles |
|
bGHR-SspIF |
249 | 0.623 ± 0.002 |
|
bGHR-SspIY |
151 | 0.378 ± 0.002 |
The results of assessing the distribution of relative occurrence rates for the SnaBI polymorphic alleles of bIGF-1 gene in the population of Herefords bred in Kazakhstan are shown in Table 7.
The data in Table 7 show that the ratio of relative occurrence rates of bIGF-1-SnaBIA and bIGF-1-SnaBIB alleles is 0.606 to 0.394, respectively. This is somewhat different from the ratio of alleles in terms of bGH-AluI and bGHR-SspI polymorphisms. Unfortunately, no data about the ratio of relative occurrence rates of bIGF-1-SnaBI polymorphism alleles in Herefords are available from other authors.
Table 7: Distribution of the relative occurrence rates of the SnaBI polymorphic alleles of bIGF-1 gene in a population of Herefords bred in Kazakhstan (Q ± SQ).
| Polymorphism | Allele | Observed occurrence rates of alleles | Relative occurrence rates of alleles |
|
bIGF-1-SnaBI |
bIGF-1-SnaBIА |
240 | 0.606 ± 0.002 |
|
bIGF-1-SnaBIВ |
156 | 0.394 ± 0.002 |
CONCLUSION
Thus, the inbreed analysis of the genetic structure of the studied Herefords populations has revealed the following.
In the population of Hereford cattle, a statistically veracious excess is observed in the occurrence rate of bGH-AluILL genotype (54.04 % of the observed occurrence rate, compared to 46.73 % of the occurrence rate theoretically calculated according to the Hardy-Weinberg’s law). Presumably, the reason for the observed deviation may be the association of bGH-AluILL genotype with the economically useful traits, which, in course of artificial selection, results in the predominance of this genotype in the population.
In terms of polymorphism of bIGF-1-SnaBI, there is an excess of the number of bIGF-1-SnaBIАВ heterozygotes observed in the population (54.55 % of the observed, compared to the theoretically expected 47.74 % according to the Hardy-Weinberg’s law). This allows assuming the association of this genotype with some breeding advantage in animal carriers.
It has been found for all three bGH-AluI, bGHR-SspI bIGF-1-SnaBI polymorphisms that in the studied Herefords populations the most widespread alleles correspond to those of other populations.
ACKNOWLEDGMENTS
This work was performed within the framework of grant-funded research project MES 2018-2020 “Integrated genetic marking of meat productivity in Hereford and Angus cattle bred in Kazakhstan by the genes that regulate growth rates” (State registration No. 0118RK00396).
Authors Contribution
All authors contributed equally.
conflict of interest
The authors declare no conflicts of interest.
REFERENCES






