Advances in Animal and Veterinary Sciences
Commentary
Prevalence of Extended Spectrum Beta-Lactamase Escherichia coli in different Cattle Feed Mills
aAgosh Verma1*, Anuradha Singh2, Suchitra Sharma3, Ruchika Atri3
1Career College, Bhopal, India; 2Associate Professor, Department of Zoology, University of Rajasthan, Jaipur, India; 3Research Scholar, Department of Zoology, University of Rajasthan, Jaipur, India.
Abstract | Feed derived pathogens are a big concern for human health as it is the beginning of the food safety chain in the farm-to-fork model. Less attention have been paid and only few studies so far have been designed to access the prevalence of these harmful bacterial colonies in animal feed. Therefore, the study was designed to assess the prevalence of ESBL-producing E. coli in cattle feed from different cattle feed mills situated in Bhopal and nearby. The objectives addressed were (1) to isolate and molecularly identify E. coli from cattle feed samples of different feed mills in Bhopal and nearby areas (2) to evaluate the antimicrobial susceptibility patterns of recovered E. coli (3) identification of genes bla CTXM grp 1,3,9, bla SHV and bla TEM in Extended-spectrum lactamase (ESBL)-producing Escherichia coli strains using PCR. A total of 200 feed samples collected which yielded 38 (19%) E.coli contaminated feed lots. Ninteen samples of feed pellets and 19 of mash feed samples were found to carrier of ESBL positive E. coli through API20E and PCR confirmation. CTXM group 2 is present in all 38 strains. E. coli isolates showed highest resistant towards drug Tetracycline and Chloramphenicol of 78.9%. On other side, they were found sensitive with Ampicillin (92.1%), Imipenem (78.9%) and third generation cephalosporins. Hygiene conditions in Feed mill industry, feed farms, trough, storage conditions, humidity, temperature, conditions in pellet machines all plays important role in E.coli contaminations. More such studies, survey and deep microbiological analysis should be promoted and is required in this field to enhance proper functioning of dairy food industries.
Keywords | Animal feed, ESBL producing E.coli, Antimicrobial susceptibility, API20E
Received | March 03, 2021; Accepted | May 18, 2021; Published | July 28, 2021
*Correspondence | Aagosh Verma, Career College, Bhopal, India; Email: draagoshverma@gmail.com
Citation | Verma A, Singh A, Sharma S, Atri R (2021). Prevalence of extended spectrum beta-lactamase Escherichia coli in different cattle feed mills. Adv. Anim. Vet. Sci. 9(9): 1302-1310.
DOI | http://dx.doi.org/10.17582/journal.aavs/2021/9.9.1302.1310
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2021 Verma et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
Food borne pathogens are the growing concern which not only affect humans but also significantly deteriorate the animal health (Health People, 2000). This illness in food producing animals can be caused by several pathogenic bacteria present in animal feed (Davis et al., 2003; Ge et al., 2013; Sharma, 2017). Animal feeds can be contaminated with foodborne bacterial pathogens (Salmonella spp., Listeria monocytogenes, E. coli, Clostiridum sp.), toxigenic fungi (genus Aspergillus and Fusarium), mycotoxins (Aflatoxins, Ochratoxin A, T-2 toxin, etc.) and pesticides (Sharma et al., 2016).
Salmonella, E.coli, Clostridium perfringens are some of the most common bacterial pathogens found in animal feed (Walker, 1959; Hacking et al., 1978; Kidd et al., 2002; Maciorowski et al., 2006; Chadwick, 2017; Davis et al., 2003; Dargatz et al., 2005; Wojdat et al., 2005, 2006; Tessari et al., 2014; Udhayavel et al., 2017). The virulent strains of these bacterial pathogens has ability to infect and colonize in the gastrointestinal tract of the animals leading to disease. Once it invades in food chain then can infect human beings also.
Pellet processing includes heat treatment and in many studies these heat-treated commercial feeds like pellets also found to contain bacterial pathogens (Carrique- Mas et al., 2007; Maciorowski et al., 2006; Sapkota et al., 2007; Cegielska – Radziejewska et al., 2013, Aury et al., 2011, Bryden, 2012). Storage, processing, transportation and environment affects the invasion of microorganisms in heat processed feed pellets and mash. Besides human to human transmission, there are various other routes through which one can get infected by E.coli that is through food chain, directly or indirectly contact with animals and also through environment (Van et al., 2019). Hence, contamination of feed with pathogenic microorganism is an important global public health issue (Crump et al., 2002; D’Mello, 2003; Walls and Buchanan, 2005; Van Immerseel et al., 2009). Therefore, microbiological evaluation of feed ingredients and processed pellet should be done to ensure further contamination (Wojdat et al., 2005).
In the EU there is a legislation (Commission Regulation (EC) No 2073, 2005) setting microbial process hygiene criterion for Enterobacteriaceae counts on foods including carcasses, milk, dairy products, and eggs. Enterobacteriaceae counts is used as a parameter determining mill hygiene and the quality of animal feeds (Veldman et al., 1995; Jones and Richardson, 2004). Many of E. coli strains reported were found to be positive for CTX-M group in there genome. There are so many factors responsible for the spread of CTX-M ESBL bacteria in animals (da Costa et al., 2007; Carattoli, 2008; Aarestrup et al., 2006; Mellmann et al., 2008) and humans, among them fecal carriage is most important (Rodrîguez – Baño et al., 2008). During the last 5 years, variety of food producing animals including cattles, chicken pigs from different countries are reported for the presence of ESBLs belonging to the CTX-M family of enzymes (Meunier et al., 2006; Madec et al., 2008). Around 82.8 % of calves in UK dairy farm were found positive for CTXM positive E.coli in 2012 (Watson et al., 2012). High rates of CTXM positive E.coli were directly related to the livestock feed, husbandry, antimicrobial usage, hygiene and waste milk (milk unfit to human consumption generally contaminated with antibiotic residues) provided to calves (Hornish and Katarski, 2002; Watson et al., 2012).
There are also scarce data on the antimicrobial susceptibility of foodborne pathogenic and indicator organisms in animal feed. In that regard, Escherichia coli are often tested as sentinel organisms due to their ubiquitous nature in animals and humans and their potential to serve as resistance gene reservoirs for human enteric pathogens (Sapkota et al., 2007; Zhao et al., 2012). Most of the E.coli strains are non pathogenic while some of them belonging to virulent categories can cause disease worldwide (Fairbrother and Nadeau, 2006).
The objective of this study and isolate pathogenic E.coli from cattle feed samples (pellet and mash) of different feed mills of Bhopal and characterize them with regard to antimicrobial resistance patterns and seasonal patterns. E.coli molecularly isolated was also identified for the presence of virulence genes bla SHV, bla TEM and blaCTX-M grp.
MATERIALS AND METHods
Sampling and Experimental Design
A pilot study of prevalence of Escherichia coli in cattle feed (mash and pellet) was done in year 2016. In this study, 200 cattle feed samples; each 100 of pellet and mash were collected from 35 different feed mills in Bhopal city. Random collection of samples was done at different time points of summer and winters. Approximately, 100 g each feed sample was collected in a sterile bag and further grounded to powder in a hygienic place. Before doing that the grinder was sanitized with 70% ethanol between different samples and allowed to dry. Samples were stored in cool and dry place until bacteriological and biochemical analyzed for Escherichia coli.
Isolation and Identification
Isolation methods used for all pathogenic E.coli are outlined in the Food and Drug Administration Bacteriological Analytical Manual (FDA - BAM) (Feng et al., 2011a). 10 gm each feed sample was enriched in 100 ml of buffered peptone water at 370C for 24 hrs. The culture was then streaked on Hichrome E.coli agar. The appearance of bluish green colonies on was considered as presumptive identification for E.coli colonies which was further confirmed by sub culturing it on eosin methylene blue agar (Producing green metallic sheen) and MacConkey agar plates (colonies are brick red in colour). These plates should be incubated at 370C for 24 hrs. The presumptive E.coli colonies were confirmed and characterized in different species by E.coli API 20E test kit (bioMérieux New Delhi, India).
Extraction of genomic DNA
The genomic DNA was extracted and the cultures which were found positive during API analysis were transferred to 2 ml Brain Heart Infusion broth (Himedia laboratories, Mumbai, India) and incubated at 37oC under constant shaking for 16 h. The bacterial cells were pelleted twice by centrifugation at 5000 rpm for 5 min and resuspended in 400 μl of SET buffer. 0.3 g Glass beads (Sigma, USA) were added to the suspension and the tubes were shaken for 90 s in a bead beater. The supernatant was then transferred to a fresh tube and combined with an equal volume of 95 phenol: chloroform: isoamylalcohol (25: 24: 1) mixed gently
Table 1: List of Primers used in the Present Study.
| Primer | Primer sequence (5’-3’) | PCR product (bp) | Annealing temperature (⁰C) |
| EC | F-CTG GAAGAGGCTAGCCTGGACGAG | 369 | 56 |
| R-AAAATCGGCACCGGTGGAGCGATC | |||
|
blaCTX-M grp-1 |
F-AAACACACGTGGAATTTAGGG | 415 | 53 |
| R-AGCTTATTCATCGCCACGTT | |||
|
blaCTX-M grp-2 |
F-CCAGCGTCAGATTTTTCAGG |
552
|
53 |
| R-CCAGCGTCAGATTTTTCAGG | |||
|
blaCTX-M grp-9 |
F-CAAAGAGAGTGCAACGGATG | 205 | 53 |
| R-ATTGGAAAGCGTTCATCACC | |||
|
blaSHV |
F-CACTCAAGGATGTATTGTG | 885 | 55 |
| R-TTAGCGTTGCCAGTGCTCG | |||
|
blaTEM |
F-TCGGGGAAATGTGCGCG | 971 | 50 |
| R-TGC TTA ATC AGT GAG GCA CC |
and centrifuged at 96 12 000 g × 2 min. The top aqueous phase was transferred to a clean tube, and one-tenth the volume, 3M sodium acetate and 2 volumes of 100% ice-cold ethanol were added. The suspension was mixed gently and stored at −20°C overnight. The sample was centrifuged at 14 000 g × 10 min, discard the supernatant and the pellet was washed with 70% ice-cold ethanol followed by centrifugation at 12 000 g × 5 min and drying pellet. The dried pellet was re- suspended in 100 μl TE buffer.
DNA quantification
The amount and quality of DNA obtained were measured using a spectramax plus 384 (Molecular devices, USA). To estimate the purity of the extracted nucleic acids, the ratio of absorbance at 260 and 280 nm (A260/A280) was used. The A260/A280 values between 1.7 and 2.0 indicate DNA samples with good quality.
Genus and species level characterization
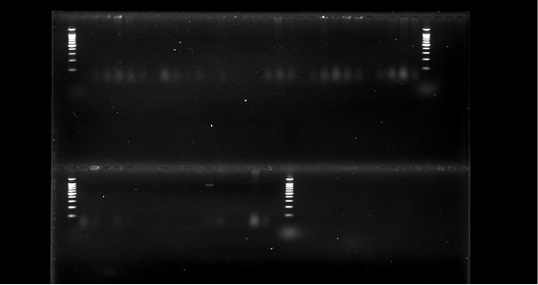
Out of total 200 cattle feed samples, 38 feedlot i.e., 19 pellet samples and 19 mash samples were found contaminated with E. coli. These phenotypically positive 38 cultures of E.coli were identified molecularly and characterize into genus and species with the help of specific primers (Figure 3) (Preethirani et al., 2015). These primers and program is elaborated in Table 1. A 25 μl PCR reaction composed of: 12.5 μl Green Master Mix (Promega, Wisconsin, USA), 0.5 μl of each primer (10 pmol), 1μl of genomic DNA (500 ng/μl) as template and nuclease free water was used for amplification of specific primers in a Bio-Rad thermal cycler (Bio-Rad Laboratories, Gurgaon, India). The amplified bands were visualized in a gel documentation system (Bio-Rad Laboratories, Gurgaon, India) after by performing gel electrophoresis.
Antibiotic sensitivity pattern
All confirmed isolates were subjected to antibiotic susceptibility testing for 11 different antibiotics. The susceptibility of obtained isolates to different antimicrobial agents was determined by disc diffusion method of (Bauer et al., 1996) using commercial available discs procured from Hi-media Laboratories, Mumbai. Antibiotic disc used were ampicillin (10μg), cefotaxime (30 μg), ceftazidime (30μg), imipenem (10 μg), naldixic acid (30 μg), ciprofloxacin (5 μg), gentamycin (10 μg), streptomycin (10 μg), co- trimoxazole (25 μg), chloromphenicol (30 μg), tetracycline (30 μg) and Nitrtrofuroin (300 mcg). The individual culture was grown at 37ºC in BHI broth until they attained a turbidity of equal to 0.5 McFarland standard. Thereafter, a sterile swab soaked in culture suspension was uniformly streaked on the surface of Mueller-Hinton agar (Hi-media laboratories, Mumbai, India) plate by rotating the plate at 60° to ensure even distribution of culture. Antibiotic discs were gently placed on the surface of the agar by using sterile forceps. The plates were next immediately incubated at 37°C for 16-18 h and zones observed were interpreted as resistance, intermediate and sensitive in accordance with Clinical and Laboratory Standards Institute (CLSI, 2015).
Molecular characterization
Isolates that were phenotypically identified as ESBL producers were screened for the presence of blaCTX-M, blaTEM and blaSHV genes by PCR using the standardized primers of other authors (Figure 4 & 5) (Pitout et al., 2010; Geser et al., 2012). Specific primers for genes blaCTX-M grp 1, blaCTX-M grp 2, blaCTX-M grp 9, blaTEM and blaSHVare presented in Table 1.
RESULTS AND DISCUSSION
Escherichia coli is the most widespread enteric bacteria, a number of strains bearing β lactamase gene can lead to illness in the animals as well as in humans. World health organization has pointed out this pathogenic group as a
key antimicrobial resistance worry. In particular, the emphasis has been given on the appearance and spread of ESBL-producing E. coli related with farm animals particularly in cattle (Liebana et al., 2012).
The hygiene and proper environmental conditions in dairy farms also plays an important role in maintaining food safety. If feed mills or cattle feed are contaminated then it will further ensure the presence of pathogens in milk that will directly affect human health. This family of the bacteria are mainly found in lower intestine of birds and mammals therefore mainly shed in feces. (Kidd et al., 2002).
In this study we have chosen some known to be good, dairy feed mills of Bhopal to trace presence of pathogenic strains of E.coli in cattle feed samples. To our research it was found out of 200 cattle feed samples collected, 38 showed presence of highly virulent β lactamase E.coli strains, which can be due to lack of adequate sanitary facilities and somehow accidently mixing of cattle feed with cattle fecal materials (da Costa et al. 2007). All 38 isolates presumed as E. coli were confirmed as E. coli after analysis by API 20 E biochemical kit (Figure 1) and validated further by PCR and found as E. coli.
Out of 38 positive samples, 10 mash samples and 11 pellet samples were isolated during months of April and May (Summer months) while 9 mash samples and 8 pellet samples were isolated from feed mills of Bhopal during January and February (Winter months). Summer prevalence is seen more that due to super shedding of cattle during summers (Figure 6) (Richard and David, 2017).
It has been seen in various studies that feed pelleting process will reduce presence of bacterial pathogens due to use of significantly high temperatures 70 oC-100 oC in the process but sometimes various other things like formulation, water content, fat content, pH also play role in killing pathogens (Hald et al., 2012; U.S. DHHS, 2018). Our results showed contamination of β lactamase E.coli in 19 mash and 19 pellet samples. After pelleting, feed transportation and storage may also interfere in contamination (Whyte et al., 2003; Maciorowski et al., 2006, Chadwick, 2017).
Molecular genetic study and Antibiotic study of these E.coli strains was done to evaluate greater picture. As, the occurrence rate of infectious diseases due to rise in antibiotic resistant E. coli has sharply increased over the past couple of years and has become a alarming issue worldwide (European Centre for Disease Prevention and Control). The Cefotaximase–München (CTX-M) enzymes are the natural β-lactamases and can be transferred between bacteria by means of plasmid transfer encoding these genes (Decousser et al., 2001). Recent reports suggests that microorganisms can be moved from animals to humans, and ESBL-producing Enterobacteriaceae bearing CTX-M and other enzymes have been earlier recovered from animals and various food products (Kornacki, 2001; Mesa et al., 2006). Therefore, it is essential to evaluate the presence, antibiotic susceptibility and ESBL associated genes in E. coli from animal feed time-to-time. Until now, scanty information is available from cattle feed in this concern from India.
In this study, the isolates were most commonly found resistant to Tetracycline, Chloramphenicol, Ciprofloxacin, Naldixic acid, Nitrofurantoin, Streptomycin and Gentamicin (Figure 2). For each of the remaining antibiotics, less than 10.0% of the strains were found as resistant. However, sensitivity to tested antibiotics was also observed. Ampicillin was found as most effective antimicrobial followed by Ceftazidime, Cefotaxime and Imipenem (Figure 2). The differences observed in the antimicrobial susceptibility patterns might be due to genetic nature of the isolate and indiscriminate use of antibiotics in veterinary and human practice. Similar findings are available where E. coli obtained from feed ingredients was found resistant to antimicrobials (Barton, 2000; Alexander et al., 2008). Earlier several authors have noticed high resistance of E. coli towards tetracycline, chloramphenicol, ciprofloxacin and nalidixic acid from feed products, and their higher use for therapeutic and prophylactic purposes in clinical science might be one plausible factor for antibiotic emergence among the strains (Johnson et al., 2003; Michalova et al., 2004; Tadesse et al., 2012).
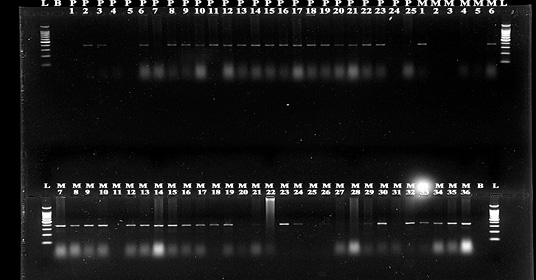
Figure 3: PCR detection of E.coli species.
Further when we performed PCR on all 38 E. coli isolates to identify the presence of CTX-M, TEM, SHV genes as the graph of ESBL producing Enterobacteriaceae is enormously increasing day-by-day at global level (Bush, 2008). These resistant bacteria are the major cause of infections related morbidity and mortality and put an unnecessary economic burden on the society (Cosgrove, 2006; Tumbarello et al., 2010). In the beginning, when these ESBL producing E. coli were isolated they were usually obtained from hospital setting, but now these bacteria are also noticed from food animals (Machado et al., 2008; Paterson et al., 2010). Increased use of different antimicrobials to combat diseases put a dominant selective pressure that may favor the emergence and dissemination of resistance genes, as suggested by some authors (Berge et al., 2006; Tragesser et al., 2006). Interestingly, 36 isolates were found positive for CTX-M group 2 that is accountable for resistance to broad-spectrum β-lactam antibiotic such as cephalosporins. Earlier studies has document that occurrence of CTX-M group 2 is rare as this group was mainly reported from Japan and South America (Canton and Conque, 2006). A previous study from India has found members of Enterobacteriaceae bears CTX-M group 1 as the main group instead of CTX-M group to and in contrast to our findings (Geser et al., 2012). The present study demonstrated that cattle feed pellets were contaminated with E. coli and are resistant to most of the commonly used antibiotics. Besides, the isolates contain the CTX-M 2 gene responsible for resistance to Beta-lactam antibiotics.
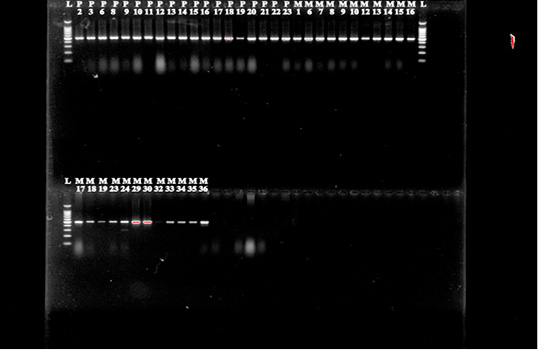
Figure 5: E.coli cultures were analyzed for the presence of blaCTX-M grp-1, blaCTX-M grp-2 and blaCTX-M grp-9 genes.
There are number of entry points for E.coli to feed, animal and human beings in contact. There should be proper hygienic conditions to avoid contamination of such bacterial pathogens. Internal working space of many feed mills are clogged by dust, molds and pests which promote to various bacterial colonies on connecting pelleting areas doors, ducts pipes etc. Moisture and humidity in feed mills and dairy farms also aids to the growth of molds, pests and bacteria in stored feed which in turn aids to contamination. Other than that, in dairy farms trough acts as a house of bacterial propagation. It infects feeding cattle, can also affect the quality of milk, also can directly or indirectly transmit infections to human beings (Jiménez et al., 2015).
Of key concern are storage of raw material, processing environment of feed mill, hygiene level of human handlers involved in the fabrication of feed, safety measures adopted during storage and transportation (Andreoletti et al., 2008). However, it is unclear the exact route of contamination of feed as the ecology of E. coli in environment is quite vast. Future studies are warranted in this direction to trace the route of transmission of E. coli in feed. Similar findings in this concern were also reported from several other authors from different parts of the world where different prevalence rate was noticed (Lynn et al., 1998; Dargatz et al., 2005; Da Costa et al., 2007; Ge et al., 2013). The differences observed in the results might be due to prevailing different hygiene levels at the processing environment, geographic location, number of samples employed for analysis and environment temperature. This preliminary study leaves a message that animal feed may act as potential source of drug resistance or pathogenic microbe in human society via food chain. This initial study provided a strong base to carry out more comprehensive large scale studies to get the better picture.
CONCLUSIONS AND RECOMMENDATIONS
AUTHORS CONTRIBUTION
Aagosh Verma: Designed the work, Collected data, Analyzed data, Drafted and Critical revision of the article.
Anuradha Singh: Designed the work.
Ruchika Sharma and Suchitra Sharma: Analyzed Data and interpretation, Drafting the Article.
Conflict Of Interest
Authors do not have any conflict of interests.
REFERENCES