Advances in Animal and Veterinary Sciences
Research Article
Co-circulation of Major Avian Respiratory Viruses in Egypt: Avian Influenza and Newcastle Disease Viruses
Ahmed Mohamed El-Sadek Hegazy1, Abeer Fathy Ibrahim Hassan2, Hala Mohamed Nabil Tolba1*
1Department of Avian and Rabbit Medicine, Faculty of Veterinary Medicine, Zagazig University, Zagazig, Egypt; 2Veterinary Hospital, Faculty of Veterinary Medicine, Zagazig University, Zagazig, Egypt.
Abstract | Avian influenza (AI) and Newcastle disease (ND) viruses are continuously affecting the Egyptian poultry industry in spite of intense vaccination schemes leading to sever economic losses. The aim of this study is to track the effect role of AIV and NDV infections in commercial layers between November 2017 and February 2019. In the present study, fifty tissue samples were collected from different layer flocks suffered from cyanosis in comb and wattles, variable respiratory manifestations and egg production problems and subjected for avian respiratory viruses screening using different diagnostic tools. Virus isolation was carried through inoculation of tissue homogenate into allantoic cavity of 9 days old SPF ECEs revealed that out of 50 samples, allantoic 35 samples were haemagglutination (HA) positive then and subjected for molecular identification based on real-time RT-PCR assay. Twenty-six allantoic fluid samples were for NDV with percentage of 74.28% while the remaining sixteen samples were positive for AIVs with percentage of 45.71% at which four samples were positive for H5 (4/16; 25%)positive, nine samples were H9 subtype positive (9/16; 56.25%) and 3 samples have mixed infection with H5 and H9 (3/16; 18.75%). Sequence analysis of the (HA and NA gene of two H5 isolates has a multi-basic amino acid motif at the cleavage site (321-PLREKRRKR/GLF-333), which is specific to highly pathogenic AIV. All H5N8 influenza isolates belonged to clade 2.3.4.4b Russian like H5N8 reassortant. The H9N2 isolates had amino acid motif at the cleavage site (333-PARSSR/GLF-341), which is specific to Low Pathogenic AIV. Furthermore, sequencing and phylogenetic analysis of M gene of selected three NDV isolates showed all related to sub genotype VIIb field strain. Our obtained results revealed the co-circulation of two major avian respiratory viruses as avian influenza and Newcastle among layers.
Keywords | Avian Influenza virus, H5N8, H9N2, Newcastle disease virus, Layer flocks
Received | September 12, 2019; Accepted | October 26, 2019; Published | December 12, 2019
*Correspondence | Hala Mohamed Nabil Tolba, Department of Avian and Rabbit Medicine, Faculty of Veterinary Medicine, Zagazig University, Zagazig, Egypt, Mohammed Ali street No.3, 44519, Zagazig, Sharkia, Egypt; Email: moonfacem2000@yahoo.com
Citation | Hegazy AME-S, Hassan AFI, Tolba HMN (2019). Co-circulation of major avian respiratory viruses in Egypt: avian influenza and newcastle disease viruses. Adv. Anim. Vet. Sci. 7(s2): 96-106.
DOI | http://dx.doi.org/10.17582/journal.aavs/2019/7.s2.96.106
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2019 Hegazy et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
Poultry industry in Egypt includes both commercial enterprises and backyard rearing (Abdelwhab et al., 2009). It has a large percentage of the supply of animal protein. Layers participate as enormous resources to the national avian flocks and this asserts the value of commercial layer flocks (Fasina et al., 2008). The rapid growth of the poultry industry in Egypt and worldwide trade as well as the live birds movement have been associated with the appearance and spread of various viral diseases (Abdelwhab et al., 2010).
Nowadays, viral respiratory diseases are a major problem in the Egyptian poultry flocks. They caused by AIV, virulent velogenic NDV and IBV (Awad et al., 2016) These pathogens, are causing disease with a huge economic impact (Roussan et al., 2008).
Outbreaks of avian influenza viruses represent a main menace to industry of poultry worldwide (Abdelwhab and Hafez, 2011). Avian influenza viruses are divided into: highly pathogenic avian influenza viruses (HPAIV) and low pathogenic avian influenza viruses (LPAIVs), according to their pathogenicity to poultry (Alexander, 2000). Highly pathogenic AI viruses cause severe respiratory manifestation and decrease in egg production with mortality up to 100% (Capua et al., 2000). While LPAI viruses induce asymptomatic manifestation to mild respiratory diseases with drop in egg production. However, it can cause high mortality in case of coinfection with other secondary bacterial pathogens (El-Zoghby et al., 2011).
In the past few years, H9N2 of Eurasian G1-like lineage has emerged into the Egyptian poultry industry as low-pathogenic avian influenza and up till now still endemic in the domesticated birds (Peacock et al., 2019). Furthermore, in 2016, H5N8 highly pathogenic avian influenza (HPAI) viruses were originally came from China; reported in Egypt with clade 2.3.4.4 group b that was related to the Eurasian HPAI H5N8 viruses (Yehia et al., 2017).
Unfortunately, the continuous and intensive use of the currently used AI vaccines could not provide the birds with sterilized immunity and stop shedding of the virus. This fact, updating the strategy for AIV control and prevention in Egypt is very critical and mandatory (Kandiel et al., 2018). The presence of HPAI H5N1 and LPAI H9N2 in Egypt influence in the epizootiologic manner to each other particularly in mixed with different application of vaccine (Arafa et al., 2012b). Since the H5N1 outbreak in Egypt in mid-February 2006, enumerous loss in the poultry industry has occurred, and the slaughter campaign overwhelmed the resources of veterinary and public health authorities (Abdelwhab and Hafez, 2011). Despite vaccination and biosecurity measures have been implemented, the disease is still endemic in Egypt and affecting the poultry and public health sectors.
Newcastle disease virus is an important viral respiratory disease to all poultry industry and a very important problem for poultry in many countries due to its effect on poultry production. Huge efforts have been made for controlling this disease. Recent studies confirmed that widely spread and circulation of NDV of genotype VII that belongs to class II, in Egypt via commercing poultry and poultry products (Elhady et al., 2018). Here in our study we investigated the current field situation of avian respiratory viruses causing real time RT-PCR assay especially for AIV subtypes H5N8, H9N2 and ND virus in Egypt. Sequence analysis were done to monitor the genetic properties of the circulating viruses in Sharkia in commercial layer flock during 2017-2019.
MATERIAL AND METHODS
Collection and preparation of samples and data
Samples from layers suffered from respiratory signs, mortalities and decrease in egg production were collected in the period from November 2017 to February 2019 from different farms in Sharkia governorate, Egypt (Table 1). The affected flocks were subjected to clinical and postmortem examination. Tissue samples (liver, lung, spleen, ovaries and oviducts) of (3-5) freshly pooled dead birds from 50 chicken layer flocks were collected. The collected tissues were pooled and homogenized in 10 mL sterile PBS to make suspension, the tissue homogenate were centrifuged at 3,000 rpm for 15 min then we collected the supernatant and transferred to sterile Eppendorf tubes containing 100 µL PEN-STREP antibiotic (Biowest company, Lot no:0510X), stored at −80 oC until used for virus isolation and real-time RT-PCR screening. The study was approved by the Committee of Animal Welfare and Research Ethics (protocol #ZU-IACU/2/F/10/2018).
Virus isolation, Antigenic Characterization and Molecular identification
Specific pathogen-free embryonated chicken eggs (SPF ECEs) were obtained from Kom Oshim, EL-Fayoum were used for virus inoculation. The tissue homogenates were inoculated into the allantoic cavity of 9–11-day-old through the allantoic route according to standard procedures (OIE, 2012). The allantoic fluid was collected from eggs with dead embryo after 24h and tested by slide Haemagglutination assay (HA). At least three successive virus passages were made for each sample to assure to be negative from avian influenza and Newcastle viruses. Viral RNA was extracted from the harvested allantoic fluids using the QIAamp viral RNA Mini kit (Qiagen, Hilden, Germany, GmbH) following the manufacturer’s instructions. Primers and probes used targeting the H5 (Londt et al., 2008) and H9 (Ben Shabat et al., 2010) subtypes and M gene of NDV (Wise et al., 2004) were used and supplied from Metabion (Planegg, Germany) Table 2.
Real time RT-PCR were carried out by adding of 25 ul to 7 ul RNA template, 12.5 ul of RT-PCR 2X probe Quanti Tect Master Mix (Qiagen, Germany), 3.625ul PCR water,50 pmol of each primer, 30 pmol of each probe and 50 pmol of Quanti Tect RT Mix. All steps at temperature 50 oC for 30 min, primary denaturation at 95 oC for 15 min and 40 cycles of denaturation at 94c for 30s. the annealing at 54c for 30 s and extension at 72 oC for 10s. The same thermal amplification of M gene for Newcastle as HA gene except annealing temperature at 55 oC for 30s. The positive specimens of H9N2, H5N8 and ND subjected to conventional RT-PCR. Extracted RNA was transcribed to CDNA by Revert Aid H Minus First Strand CDNA
Table 1: Descriptive data of examined layers suspected to be infected with AIV and NDV within Sharkia province (2017-2019).
| Locality | Vaccination with H5 and H9 vaccine | Effect on egg production | Severity of respiratory signs | Mortality/ daily | Breed | Age/week A | Birds no | Flock no fl |
| SHIBA |
-VE - |
↓10% | +++ | 10 | Lohman elwadi | 20 | 1000 | 1 |
| Qinaiat | -ve | ↓1-2% | + | 10 | H and N | 16 | 1000 | 2 |
| Hehya | +ve (H5 vaccine) | ↓5-10% | ++ | 15-20 | Lohman elwadi | 30 | 2350 | 3 |
| Hehya | +ve (H5 vaccine | ↓5-10% | ++ | 15-20 | Isa brown | 30 | 2850 | 4 |
| Tal Raq | +ve | ↓20% | +++ | 20-70 | Bovans | 30 | 5500 | 5 |
| Fakous | +ve | ↓20-25% | ++ | 15 | TITRA | 24 | 1000 | 6 |
| Fakous | +ve | ↓from 91% to 42% | +++ | 500 | Lohman elwadi | 25 | 12000 | 7 |
| Ibrahimia | -VE | ↓10% | ++ |
10-15, 10-1 |
H and N | 24 | 1500 | 8 |
| Abo Kabir A | +ve | Not affected | + | 20-25 | Lohman elwadi | 32 | 4000 | 9 |
| Abo Kabir | +ve | ↓10% | + | 25 | Bovans | 40 | 4000 | 10 |
| Bilbis | +ve | ↓15% | ++ | 1-2 | H and N | 30 | 8000 | 11 |
| Kafr Sakr | +ve | ↓15-20% | ++ | 20-25 | Bovans | 28 | 3000 | 12 |
| Hehya | +ve | ↓40%within 20 days | +++ | 10 | Lohman elwadi | 30 | 9500 | 13 |
| Kafr Sakr | +ve | ↓25% | ++ | 30 | H and n | 28 | 6000 | 14 |
| Shobak S | +ve | ↓15% | + | 10-15 | lohman | 31 |
250025 |
15 |
| Diarb Negm D | +ve | ↓10% | ++ | 20 | Lohman elwadi | 28 | 5000 | 16 |
| Ibrahimia | +ve | ↓25% | +++ | 15-20 | TITRA | 30 | 2500 | 17 |
| Fakous | +ve | ↓20% | +++ | 50 | H and N | 28 | 7000 | 18 |
| Hehya | +ve | ↓15% | ++ | 10-15 | Bovans | 29 | 3000 | 19 |
| Hehya | +ve | ↓2-3% | + | 20 | Lohman elwadi | 29 | 10000 | 20 |
| Elsalhia diarb Ne | +ve | ↓↓5-10% | +++ | 25 | H and N | 33 | 5500 | 21 |
| Diarb Negm | +ve | ↓5% | ++ | 10-15 | Lohman elwadi | 31 | 2000 | 22 |
| Fakous | +ve | ↓15% | ++ | 20 |
Isa brown |
20 | 1000 | 23 |
| qinaiat | +ve | ↓25% | ++ | 20 | H and N | 32 | 2500 | 24 |
| Diarb Negm | +ve | ↓10% | ++ | 15-20 | Lohman elwadi | 40 | 4000 | 25 |
| Kafr Sakr | +ve | ↓3-5% | ++ | 15-20 | Lohman elwadi | 35 | 5000 | 26 |
| Shirwida | +ve | ↓15% | +++ | 30-35 | Bovans | 33 | 4000 | 27 |
| Fakous | +ve | ↓10% | ++ | 25 | H and N | 29 | 1500 | 28 |
| Ibrahimia | +ve | ↓10% | ++ | 10-15 | Bovans | 30 | 1500 | 29 |
| Ibrahimia | +ve | ↓1-2% | ++ | 30-35 | Lohman elwadi | 30 | 8000 | 30 |
| Hehya | +ve | Not affected | + | 15 |
Is a brown |
29 | 4000 | 31 |
| +ve | ↓5-10 | ++ | 20-25 | Lohman elwadi | 35 | 2000 | 32 | |
| Diarb Negm | +ve | ↓35% | ++ | 10 | H and N | 28 | 3000 | 33 |
| Fakous | +ve | ↓20% | ++ | 25-30 | Lohman elwadi | 30 | 5500 | 34 |
| Banayos | +ve | ↓3-5% | ++ | 25 | Bovans | 29 | 1000 | 35 |
| Met abo ali | +ve | ↓15% | ++ | 50 | H and N | 30 | 3550 | 36 |
| Hehya | +ve | ↓10% | ++ | 15-20 | Bovans | 29 | 2700 | 37 |
| Fakous | +ve | ↓10% | ++ | 25 | H and N | 27 | 2000 | 38 |
| Borden BO | +ve | ↓5% | ++ | 20 | Lohman elwadi | 33 | 1500 | 39 |
| Ibrahimia | +ve | ↓2-3% | ++ | 25 | Lohman elwadi | 30 | 4000 | 40 |
| Ibrahimia | +ve | ↓15% | ++ | 10 | H and N | 29 | 3000 | 41 |
| Diarb Negm | +ve | ↓5% | ++ | 5 | Bovans | 30 | 2500 | 42 |
| Hehya | +ve | ↓5% | ++ | 10-15 | H and N | 35 | 6000 | 43 |
| Ibrahimia | +ve | ↓10% | ++ | 5 | Lohman elwadi | 48 | 4000 | 44 |
| Kafr Sakr | +ve | ↓20% | ++ | 10 | Bovans | 29 | 2000 | 45 |
| banayos | +ve | ↓10% | ++ | 25 | Lohman elwadi | 30 | 2000 | 46 |
| Fakous | +ve | ↓25% | ++ | 20 | H and N | 33 | 9000 | 47 |
| Hehya | +ve | ↓15% | ++ | 50-70 | Lohman elwadi | 30 | 5000 | 48 |
| Shobak | +ve | ↓10% | ++ | 15 | H and N | 28 | 2000 | 49 |
| Abo Kabir | +ve | ↓10% | ++ | 25 | Bovans | 40 | 10000 | 50 |
Table 2: Primers and probes used for real time PCR.
| Virus | Gene | Primer/ probe sequence 5'-3' | Ref |
| AI | M |
Sep1 AGATGAGTCTTCTAA CCGAGGTCG |
|
|
Sep 2 TGCAAAAACATCTTC AAGTCTCTG |
|||
|
SEPRO [FAM]TCAGGCCCC CTCAAAGCCGA [TAMRA] |
|||
| H5 |
H5LH1 ACATATGACTAC CCACARTATTCA G |
||
|
H5RH1 AGACCAGCT AYC ATGATTGC |
|||
|
H5PRO [FAM]TCWACA GTGGCGAGT TCCCTAGCA[TAMRA] |
|||
| H9 |
H9F GGAAGAATTAATTATTATTGGTCGGTAC |
||
|
H9R GCCACCTTTTTCAGTCTGACATT |
|||
|
H9 Probe [FAM]AACCAGGCCAGACATTGCGAGTAAGATCC[BHQ] |
|||
| ND | Matrix |
M+4100 AGTGATGTGCTCGGACCTTC-3’ |
|
| M-4220 CCTGAGGAGAGGCATTTGCTA-3’ | |||
| M+4169 [FAM]TTCTCTAGCAGTGGGACAGCCTGC[TAMRA]-3’ |
Table 3: Primers used in Reverse Transcriptase-Polymerase Chain Reaction (one step RT-PCR) and Sequence reaction of HA and NA genes of H9N2.
| Prime ID | Primer Sequence for HA gene amplification | Reference |
| F1-6 | 5’ TAG CAA AAG CAG GGG AAT TTC TT 3’ | RLQP |
| H9- Rev | 5’ GCC ACC TTT TTC AGT CTG ACA TT 3’ | |
| H9-For | 5’GGA AGA ATT AAT TAT TAT TGG TCG GTA C 3’ | |
| HT7R | 5’ TAA TAC GAC TCA CTA TAA GTA CAA ACA AGG GTG 3’ | SEPRL |
| Primer Sequence for NA gene amplification | ||
| N2-R950 | CGC CAA CAA GTC CTG AGC ACA CAT | RLQP |
| N2-F630 | CAT GGG ATG CTT ACC GAC AGT ATT | RLQP |
Table 4: Primers used in Reverse Transcriptase-Polymerase Chain Reaction (one step RT-PCR) and Sequence reaction of HA and NA genes of H5N8.
| Prime ID | Primer Sequence for HA gene amplification | Reference |
| HGGT |
5’ CTC TTC GAG CAA AAG CAG GGG T 3’ |
RLQP |
| KH3 | 5’ TAC CAA CCG TCT ACC ATK CCYTG 3’ | |
| H5F5-1088 | 5’ TTG GAG CTA TAG CAG GTT TTA TAG AGG 3’ | |
| Bm-NS-890R | 5’ ATA TCG TCT CGT ATT AGT AGG AAA CAA GGG TGT TTT 3’ | SEPRL |
| Primer Sequence for NA gene amplification | ||
| f1- N8 |
5’GCA AAA GCA GGA GTT TAA AAT GAA TCC 3’ |
RLQP |
| R778-N8 |
GCC TTG ATT TGC TTT GT 3’ 5’ |
RLQP |
Synthesis Kit Fermentas Inc., Walthan, MA, USA, according to manufacturer instructions Tables 3 and 4.
Sequencing, Sequence analysis and phylogenetic analysis
we select Two H5N8, three H9N2 and three ND isolates as they cause high mortalities and severe decrease in egg production for sequencing by using Bigdye Terminator V3.1 cycle sequencing Kits (Perkin-Elmer, Foster city, USA). HA and NA subtypes for AIVS known by nucleotide BLAST (http://www.ncbi.nim.nih.gov/BLAST) and recorded in Gene Bank with accession numbers for HA were MK975994, MK975995, MK968882, MK968881 and MK968880 and for NA were MK975996, MK975997, MK968894, MK968893 and MK968892. The phylogenetic tree was performed by neighbor-Joining method in MEGA version 7 (http://www.megasoftware.net). The tree topology was evaluated by 1,000 bootstrap analyses.
Results
Clinical and postmortem examination
In the present study layers showed, comb and wattle edema with cyanosis, respiratory signs, greenish watery diarrhea, shell-less egg, and soft eggs were reported with drop in egg production percentage with 10-15% mortality were also recorded. Postmortem examination revealed congested trachea, pneumonic lung, air sacculitis. There was petechial hemorrhage in proventriculus and pectoral muscle, hemorrhagic enteritis, egg peritonitis and hemorrhage in ovarian follicle (Figure 1).
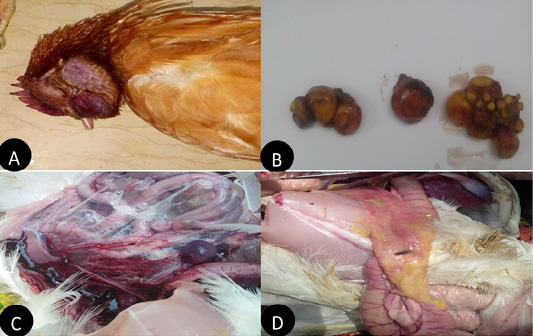
Figure 1: Clinical manifestations and PM lesions of laying chickens suspected to be infected with AIVs and NDV. (A) severe congestion in comb, wattle and face edema in 28week flock No.13. Belbais; (B) severe hemorrhage and congestion of ovary; (C) severe congestion in spleen and ascites in 28 week flock No.19. Hehia; (D) hemorrhage and congestion in duodenum, pancreas in 25 week flock No.5 Talraq.
Virus isolation, Haemagglutination and Molecular identification
Inoculation of embryonated chicken eggs with tissue homogenates from bird samples showed mortality and AIV and NDV lesions in embryos after 24 h in all samples. Haemagglutinating viruses were detected in 35 samples (70%) as in (Table 5). Positive HA allantoic fluids from samples were subjected to real-time RT-PCR to identify NDV and AIVs subtypes (H5N8 and H9N2). The results revealed that 26 (74.28%) out of 35 were positive NDV genotype VII field isolate and 16 (45.71%) out of 35 were positive for AIVs, out of 16 positive AIVs 4 flocks (25%) were H5 subtype positive 9 flocks (56.25%) were H9 subtype positive and 3 (18.75%) mixed flocks with H5 and H9. All the samples obtained from vaccinated flocks against both Newcastle and influenza viruses. The Ct values from examined samples are in (Table 5).
Table 5: Result of Haemagglutination assay and CT value of Real time RT PCR.
|
CT |
Slide HA | Code no | Flocl no | ||
| ND | H9 | H5 | |||
| 16 | NO CT | NO CT | + | 5 | 1 |
| 15 | 19 | 26 | + | 7 | 2 |
| 17 | NO CT | 14 | + | 13 | 3 |
| 27 | 13 | NO CT | + | 14 | 4 |
| 16 | 25 | NO CT | + | 15 | 5 |
| 26 | 11 | 17 | + | 16 | 6 |
| 31 | NO CT | NO CT | + | 18 | 7 |
| 23 | NO CT | NO CT | + | 19 | 8 |
| 26 | NO CT | NO CT | + | 20 | 9 |
| 11 | NO CT | NO CT | + | 21 | 10 |
| 32 | NO CT | NO CT | + | 22 | 11 |
| 26 | NO CT | NO CT | + | 23 | 12 |
| 26 | NO CT | NO CT | + | 24 | 13 |
| 22 | 15 | NO CT | + | 26 | 14 |
| 14 | NO CT | 14 | + | 30 | 15 |
| 14 | 26 | NO CT | + | 31 | 16 |
| 14 | NO CT | NO CT | + | 32 | 17 |
| 27 | NO CT | NO CT | + | 33 | 18 |
| 18 | NO CT | NO CT | + | 34 | 19 |
| 26 | 14 | NO CT | + | 35 | 20 |
| 22 | 26 | 29 | + | 36 | 21 |
| 16 | NO CT | 17 | + | 40 | 22 |
| 24 | 14 | NO CT | + | 41 | 23 |
| 24 | 26 | NO CT | + | 42 | 24 |
| 23 | NO CT | 28 | + | 49 | 25 |
| 22 | 26 | NO CT | + | 50 | 26 |
| -VE | -VE | -VE | + | 8 | 27 |
| -VE | -VE | -VE | + | 9 | 28 |
| -VE | -VE | -VE | + | 44 | 29 |
| -VE | -VE | -VE | + | 47 | 30 |
| -VE | -VE | -VE | + | 48 | 31 |
| -VE | -VE | -VE | + | 39 | 32 |
| -VE | -VE | -VE | + | 28 | 33 |
| -VE | -VE | -VE | + | 10 | 34 |
| -VE | -VE | -VE | + | 6 | 35 |
Sequencing and phylogenetic analysis
H5N8, H9N2 and NDV isolates from laying hens were confirmed further by sequencing. The phylogenetic analysis of HA gene sequences (Figure 2) and NA gene sequence (Figure 3) of the three Egyptian isolates (H9N2; A/chicken/Egypt/AB2/2018, A/chicken/Egypt/AB3/2018 and A/chicken/Egypt/AB4/2018) showed that they were closely related to the other Middle East H9N2 strains. Our isolates shared the common ancestor with A/Qa/HK/G1/97 present in Asia of one group (Egy/G1) related to the G1 lineage within group B. (Abdelhafez et al., 2019) Sequencing of the HA segment revealed amino acid motif at the cleavage site (333-PARSSR/GLF-341), which is characteristic of LPAIV. When comparing the three isolates togeather, the NA gene showed 100% identity whereas the HA showed approximately 99% amino acid identity. as in (Figure 7).
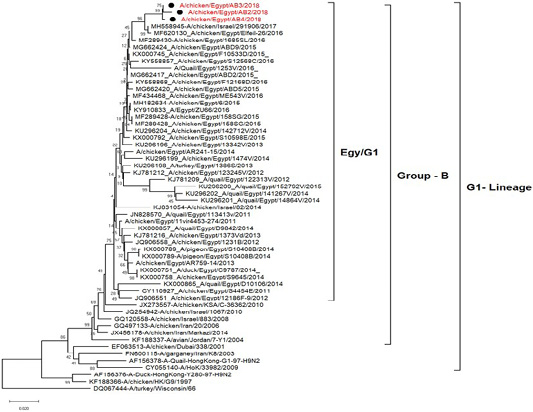
Figure 2: Phylogenetic tree of amino acid sequence of the hemagglutinin genes of three field isolate of avian influenza subtype H9N2 (black circle) viruses isolated in Egypt during 2017–2019 and with reference strains from GenBank.
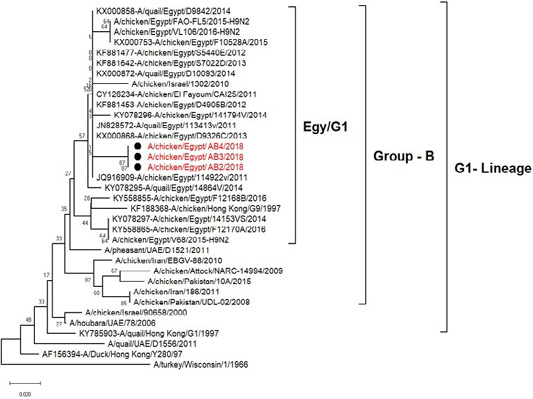
Figure 3: Phylogenic tree of amino acid sequence of NA gene of three field isolate subtype H9N2 (black circle) with other reference strains in gene bank.
The phylogenetic analysis of HA gene sequences and NA gene sequence (Figures 4 and 5) of our Egyptian two isolate (H5N8) showed that our isolates belonged to Russian like H5N8 reassortant which was named A/chicken/Egypt/AB1/2018 and A/chicken/Egypt/AB2/2018. Sequencing of HA gene of the two isolates showed a multi-basic amino acid motif at the cleavage site (PLREKRRKR/GLF/), which is characteristic to HPAIV. They were compared with other H5N8 isolates and Avian influenza vaccines used commercially on the gene bank. The results showed that the two isolates present in one group with Russian strains and belonged to clade 2.3.4.4b, this indicated that the Russian HPAI H5N8 virus from Russia, Europe (A/great-creste_grebe/Uvs-Nuur_Lake/341/2016) is the origin of the reassortant Egyptian H5N8 viruses. No mutation and gross deletions was detected among the two Egyptian H5N8 viruses. The NA and HA gene of our isolates showed amino acid identity with other selected isolate with percentage of 84.7-100% as in (Figure 7).
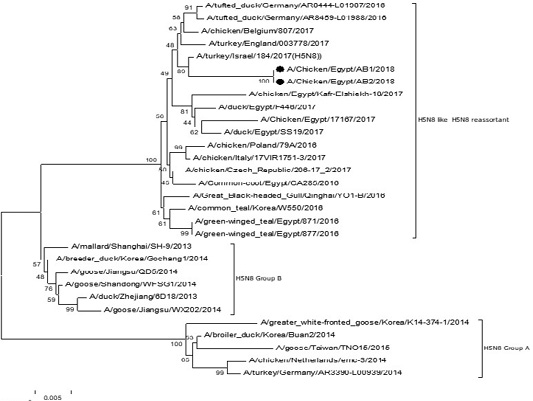
Figure 4: Phylogenic tree of amino acid sequence of HA gene of two field isolates highly pathogenicH5N8 (black star) with other reference strains in gene bank
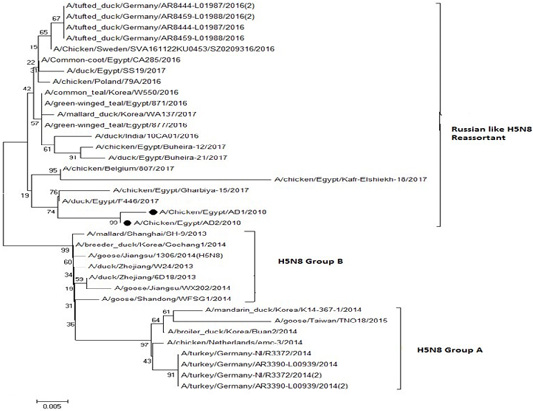
Figure 5: Phylogenic tree of amino acid sequence of NA gene of two field isolate subtype H5N8 (black circle) with other reference strains in gene bank.
Partial sequences of the selected isolated strains of NDV for M gene phylogenetically analyzed and showed that they belong to genotype VII vNDV (Figure 6). The vNDV isolates in our study were 100% typical to each other based on amino acid and nucleotide identities. Compared to other recent Egyptian strains isolated during 2011–2016, the identity ranged between 94.1–99.6% and 96.7– 100% on nucleotide and amino acid levels, respectively.
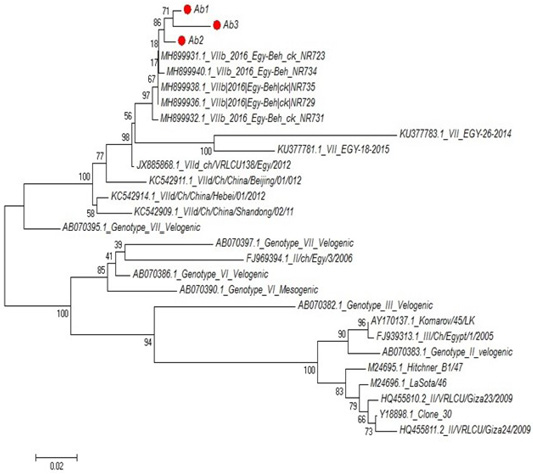
Figure 6: Phylogenetic tree of selected field NDV (3 isolates) indicated by red circle with other related reference strains of NDV in gene bank.
Discussion
Layers shared enormous resources to the avian flock and this asserts the importance of layer flocks (Fasina et al., 2008), also the average mortality percent was the minimal. This may be as outcome of cage housing which prevent the movement and contact between infected and non-infected bird while in broilers reared in floor raised system and share water and feed sources. Not withstanding respiratory disease outbreaks have been in increase in layer flocks that caused by mainly by AIV, virulent NDv or IBv (Awad et al., 2016). These pathogens have a high significance and a big economic impact as they can cause diseases alone or mixed with each other (Roussan et al., 2008).
In the current study clinical examination of layers naturally layer flocks suspected to be infected with respiratory viruses revealed cyanosis in comb and wattle as well as facial edema, respiratory signs, greenish watery diarrhea and egg structural defects with drop in egg production. Postmortem examinations revealed congested trachea, pneumonic lung, air sacculitis and ovarian follicular hemorrhages. In Nigeria (Adene et al., 2006) cleared that laying hens naturally infected with AIV showed respiratory and intestinal lesions and 20% out of 248 laying hens showed nervous signs in young age. Also, other researchers confirmed the presence of neurological signs with no specific reference to age (Joannis et al., 2006) but Guan and colleagues (Guan et al., 2000) showed that coughing, sneezing and decline in production of egg with low percent is characteristic for H9N2 infection. The majority of AIV positive flocks were affected with H9 subtype with percentage 56.25%. Also, Francesco Bonfante et al. (2018) reported that H9N2 virus as a primary pathogen in layer hens, and is responsible for mild respiratory signs and drop of egg production with severe salpingitis. Infection of birds with LPAI viruses have no or few clinical signs (Bertran et al., 2014). Also, it may be transformed into pathogenic virus by way of mutation (OIE, 2008).
In the current study AIV subtypes H5N8 and H9N2 and NDV were investigated in commercial layer flocks in Egypt during the period of November 2017 to February 2019. All samples were isolated in SPF-ECEs though allantoic sac route and make death to the embryos with hemorrhagic lesions (Salaheldin et al., 2018). Also, our study showed that Haemagglutinating viruses were detected in 35 samples (70%) out of 50 flock samples. Positive harvested allantoic fluids were investigated by real-time (RT-PCR for detection NDV and AIVs subtypes (H5N8 and H9N2) as it represents accurate and sensitive method for AI detection (Bouwstra et al., 2015). The results revealed that 26 (74.28%) out of 35 were positive NDV genotype VIIb field isolate and 16 (45.71%) out of 35 were positive for AIVs. Out of 16 nine flocks (56.25%) were H9 subtype positive, four flocks (25%) were H5 subtype positive and 3 (18.75%) mixed flocks with H5 and H9. All the samples were from vaccinated flocks against Newcastle and influenza viruses. Our results were in accordance with those of (Arafat et al., 2018) they cleared that, the highest detection results for respiratory viruses were NDv (62.2%) followed by AI H9 (58%) then AI H5 virus (17.5%). Also (Samy and Naguib, 2018) reported that, in chickens Avian Influenza is one of the main causes of diseases affecting respiratory system with economic importance worldwide. In our study the mixed infection results observed only in three flocks where the H9N2, ND and H5N8 were isolated, this type of infection is very dangerous and causes high mortality to the infected laying birds. The causes of co- infection attributed to the infection with some viruses considered as a stress to the birds and facilitated the infection with other viruses. (Watanabe et al., 2018). The mixed infection with two subtypes of avian influenza has resulted in virus reassortment which associated with both increase in mortality and the dispersal of infection between poultry (Kayali et al., 2014). Egypt is believed to be a hotspot for the generation of new subtypes (Abdelwhab and Abdel-Moneim, 2015). Real time RT/PCR performing and then sequencing following by phylogenetic analysis is very remarkable and worthy protocol to identify subtypes of Avian influenza (Salaheldin et al., 2018).
In our study Two positive samples of PCR to (H5) were sequenced. The cleavage site of amino acids (PLREKRRKR/GLF) indicate that the two isolates are highly pathogenic Avian influenza virus as reported by (Kandeil et al., 2017) The HA and NA gene phylogenetic analyses showed that the two Egyptian HPAI H5N8 viruses clustered with 2.3.4.4b viruses from Russia, Europe, and Asia. Our results agreed with (Yehia et al., 2017) who registered the incursion of HPAI H5N8 virus of clade 2.3.4.4b and (Salaheldin et al., 2018). The present new subtypes need conciderations in vaccination prepare for good control and prevention. Also the three isolates of H9N2 have the same motif of low pathogenic avian influenza virus (PARSSR/GLF) as reported by (Steinhauer, 1999) and placed within G1B, the same lineage circulating in the Middle East as previously shown (Arafa et al., 2012). This motif has secreted only in respiratory organs and intestine (Neumann and Kawaoka, 2006) The incidence of H9N2 infections was low in layers and breeders. (Soliman et al., 2014). In previous studies HA gene was classified phylogeneticaly into 2 sub-lineages of group B (Kandeil et al., 2014). The proteolytic cleavage site on HA is the main pathogenicity factor of influenza viruses also the co- infection of NDV with H9 subtypes could be a factor in increased the severity of AI infection. The reason for that may be due to the cleavage activation of the HA. The cleavage of the HA play a key role in viral pathogenicity (El-Zoghby et al., 2011) The partial sequence analysis of HA and NA revealed that HA genes share similarity 100% with each other and NA 99% with other strains, the first Egyptian quail isolate in 2011 (Arafa et al., 2012)
The phylogenetic analysis of partial sequences of the selected isolated NDV strains for M gene showed that they belong to genotype VII velogenic strain. The amino acid sequences of the M protein proteolytic cleavage site motifs (residues 10 to 120) of the NDv strains M-gene were compared. The vNDV isolates in this study were 100% typical to each other based on amino acid identity. Compared to other recent Egyptian strains isolated during 2011–2016, the identity ranged between 90.6%–97.3% homology between each other, homology with field strain MH899832-1-2016-Egy-beh-chNR731(97%:98.4%) and homology with vaccinal strain Y18699Colone 30 is (77.5%: 79.1%). These results in accordance with (Awad et al., 2015).
CONCLUSIONS
In our study many respiratory viral diseases were recorded which make a threat to poultry industry in Egypt. NDV virus and H9N2, H5N8 avian influenza subtypes isolated from different localities in Egypt plus the high distribution of avian influenza H5N1among layer flocks in 2017 and 2019. Birds in ranches are stores assuming a role in the spread of the infection and delivering a general wellbeing hazard. Proper clean measures ought to be connected on ranches to control the presentation of birds and in this manner people to the wellspring of infection. Proceeds with surveillance and observing of the circulating viruses is significant for understanding the development of the viruses and to all the more likely selected viruses for immunization concentrates to limit the wide spread of the viral infection.
ACKNOWLEDGEMENTS
The authors are thankful to Prof Dr. Fatma Professor of Virology and Vet., Faculty of Veterinary Medicine, Zagazig University for her valuable advice in analyzing the sequencing data.
Authors Contribution
Ahmed Mohamed EL-Sadek Hegazy provided guidance, technical support and edited the manuscript and reviewed drafts of the paper, approved the final draft. Abeer Fathy Ibrahim Hassan prepared figures and/or tables. Hala Mohamed Nabil Tolba analysed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest
REFERENCES







