Advances in Animal and Veterinary Sciences
Research Article
Overview on the Most Prevailing Bacterial Diseases Infecting Oreochromis niloticus at Aswan Fish Hatchery, Egypt
Awatef Hamed Hamouda1*, Eman Moustafa Moustafa2, Mohamed Mamdouh Zayed3
1Fish Diseases Department, Faculty of Fish and Fisheries Technology, Aswan University, Aswan, Egypt; 2Department of Fish Diseases and Management, Faculty of Veterinary Medicine, Kafrelsheikh University, Kafr El-Sheikh, Egypt; 3Department of aquaculture, Faculty of Aquatic and Fisheries Sciences, Kafr El-Sheikh University, Egypt.
Abstract | Information about bacterial diseases from aquaculture systems at Aswan Governorate, Egypt is scarce and old, making the interpretation, prevention and treatment of their episodes very difficult. This study aimed to shed light on the most predominant bacterial infections in Oreochromis niloticus from Sahary fish hatchery; with special focus on isolation, identification, and characterization of pathogenic bacteria in this cultured fish. During an epizootic; from April to May 2018, one hundred clinically diseased Oreochromis niloticus were collected alive from the hatchery. Diseased fish showed hemorrhagic patches distributed all over the body, detachment of scales, fin erosions, distended abdomen and corneal opacity with exophthalmia in some cases. Collected aseptic tissue samples were cultured over different selective differential media for phenotypic characterization. Moreover, experimental infection using the most prevalent pathogenic bacteria (Aeromonas hydrophila) was conducted. Biochemical tests, revealed the presence of Aeromonas hydrophila, Aeromonas sobria, Pseudomonas fluorescens, Enterococcus faecalis, Staphylococcus aureus and Streptococcus agalactiae with prevalence of 56%, 45%, 33%, 11%, 13% and 14%, respectively. Molecular identification, antimicrobial susceptibility, multiple antibiotic resistance (MAR index) and histopathological changes induced by Aeromonas hydrophila were investigated. Aeromonas hydrophila molecularly revealed an amplified product of 356 bp size using the 16S rRNA gene and its pathogenicity is confirmed by both infectivity experiments and histopathological examination. Aeromonas hydrophila is sensitive to Gentamicin, Ciprofloxacin, Amikacin and Nalidixic acid recording average MAR index of 0.534 which has been indicative of isolates associated with a high risk of proceeding from environments with usual antimicrobials use. Aeromonas hydrophila is completely resistant to Penicillin and Erythromycin.
Keywords | Antimicrobial resistance, Experimental challenge, Histopathological alteration, Molecular characterization, Tilapia, Pathogenic bacteria.
Received | June 25, 2019; Accepted | August 19, 2019; Published | October 15, 2019
*Correspondence | Awatef Hamed Hamouda, Fish Diseases Department, Faculty of Fish and Fisheries Technology, Aswan University, Aswan, Egypt; Email: awatefhamouda@aswu.edu.eg; awatefhamouda@yahoo.com
Citation | Hamouda AH, Moustafa EM, Zayed MM (2019). Overview on the most prevailing bacterial diseases infecting oreochromis niloticus at aswan fish hatchery, egypt. Adv. Anim. Vet. Sci. 7(11): 950-961.
DOI | http://dx.doi.org/10.17582/journal.aavs/2019/7.11.950.961
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2019 Hamouda et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Tilapia is an ideal candidate for aquaculture especially in developing countries. It has a fast growth rate, able to reproduce in captivity, has a short generation time, resist stress and diseases, accept artificial feeds immediately after yolk-sac absorption, feed on low trophic levels and tolerate a wide range of environmental conditions (El-Sayed, 2006). Oreochromis niloticus, the most important aquaculture species of the 21st century (FAO, 2012); contributes
about 83% of the total global tilapia production (Gupta and Acosta, 2004; Bostock et al., 2010 ; Gaafar et al., 2015). It is considered as the most common and popular fish in Egypt, and has proved to be vital, contributing from 30 to 40 % of the total production of the Lake Nasser in Egypt (Latif and Rashed, 1983). However, tilapia cultivation may display some health problems, which are associated with the presence of pathogenic bacteria (Pech et al., 2017).
Fish diseases constitute one of the most important challenges confronting fish cultivation especially the bacterial ones. Bacterial fish pathogens are naturally present in the fish surrounding environment; however, under certain stress conditions, may cause severe economic losses with 80% of mortalities in fish farms (Austin and Austin, 2007). Bacterial diseases are the most common diseases in intensive fish rearing facilities which could hinder the aquaculture industry (Ibrahim et al., 2013). It may be the most significant cause of economic loss and sometimes have zoonotic threats to human consumers (Plant and LaPatra, 2011). The list of new pathogenic bacterial species isolated from the fish has been quickly expanding coordinating with the fast development and intensification of aquaculture, contamination and expanded utilization of water bodies (Harvell et al., 1999).
Aeromonas hydrophila may cause septic arthritis, diarrhea, gastroenteritis, skin and wound infections, meningitis, septicemia, otitis media, endocarditis, peritonitis, cholecystitis, hemolytic uremic syndrome and food poisoning (Ko et al., 2000; Tsai et al., 2006; Guerra et al., 2007; Salunke et al., 2015). Some strains of Pseudomonas fluorescens found to be opportunistic human pathogens (Younes and Gaafar, 2014). Staphylococcus aureus is a major human pathogen (Edwards et al., 2012; Soliman et al., 2016). The infection of Streptococcus agalactiae has caused neonatal meningitis in human and mastitis in cows (Bohnsack et al., 2004; Lindah et al., 2005). Among bacteria that cause mortalities in Oreochromis niloticus, pathogens of Aeromonas sp., Pseudomonas sp., Enterococcus sp., Staphylococcus sp. and Streptococcus sp. could be reported (Ahmed and Shoreit, 2001; El-Ashram, 2002; Carnevia et al., 2010; Younes and Gaafar, 2014; Liu et al., 2019).
This study aims to isolate, identify, and characterize pathogenic bacteria of cultured Oreochromis niloticus in Sahary fish hatchery, Aswan Governorate, with special emphasis on molecular identification, pathogenicity, antimicrobial susceptibility, multiple antibiotic resistance (MAR index) and histopathological changes induced by the most prevalent isolated pathogenic bacteria. The information generated will be a starting point for pathogenic bacteria prevention and control plans in this region.
Material and methods
This study was conducted at Sahary fish hatchery in Aswan Governorate, Egypt which is a government fish hatchery constructed to aid and enhances the development of overfished Lake Nasser fishery by annually release of Oreochromis niloticus seed to support the wild stock in the natural water body. In 2016, the hatchery produced 7 million fry (GAFRD, 2016) and in 2018, it produced 10 million and 395 thousand fry.
Material
Naturally infected Fish: One hundred clinically diseased Oreochromis niloticus were collected alive from the hatchery in April and May 2018 with different body weight (30-550 g) and total lengths (10-35cm) during an epizootic with septicemic clinical signs and mortality rates. The collected fish were transferred to the wet Lab., Fish Diseases Department, Faculty of Fish and Fisheries Technology, Aswan University, Egypt and held in well-prepared glass aquaria supplied with sufficient amounts of dechlorinated tap water with continuous aeration using an electric air pump (Eissa, 2016) till investigation.
Fish for Pathogenicity Test: A total number of 150 apparently healthy Oreochromis niloticus weighing (50±5 g) obtained from the same hatchery and transported to the wet Lab. of Fish Diseases Department at Faculty of Fish and Fisheries Technology. The fish were kept for 15 days in well prepared glass aquaria before the beginning of the experimental infection for acclimatization and for examinations of random samples to insure their clearance of any natural infection.
B- Methods
Clinical and postmortem examinations:
Oreochromis niloticus were clinically examined to record any apparent clinical abnormalities and postmortem lesions according to Austin and Austin (2007) and Eissa (2016) in accordance with the protocol No. (1/2018) approved by Faculty of Fish and Fisheries Technology, Aswan University, Egypt.
Bacterial Isolation: Tissue samples were aseptically collected from skin, liver, kidney and spleen of the examined fish, pre-enriched on Tryptic Soy Broth and incubated for 24 hrs at 28˚C. After that, they were streaked on tryptic soy agar (TSA) and incubated for 48 hrs at 28˚C. Pure single colonies on TSA were further streaked on selective media; Rimler Shotts agar (RS), Aeromonas selective agar base with Ampicillin supplement, Pseudomonas-F-agar base medium, XLD medium, Mannitol Salt Agar (MSA), Triple sugar iron agar (TSI), MacConkey agar plates, blood agar and Streptococcus selective agar and incubated for 24 hrs at 28-37 °C. Moreover, tissue samples were selectively pre-enriched on Enterococcus M broth medium and incubated for 18-24 hrs at 37°C and then streaked on BBLTM Enterococcosel Agar (EA) and incubated overnight at 37°C; especially for isolation of Enterococci bacteria. Bacterial colonies were classified according to Reynolds (2011).
Phenotypic identification of bacterial isolates: Phenotypic identification of the bacterial isolates was conducted as previously described (Schäperclaus et al., 1992; Bergey’s, 1994). Gram staining and motility test were performed (Cruickshank et al., 1975). All isolates were identified biochemically following the criteria proposed by Kreig and Holt (1984) and MacFaddin (2000).
Molecular identification by Polymerase Chain Reaction (PCR): The most prevalent bacterial isolate was molecularly identified using PCR.
Primer sequences used for PCR identification system:
The primer used for detection of Aeromonas hydrophila is A16S1 (F)-A16S1(R) (Pharmacia Biotech) (A16S1F 5′ GGGAGTGCCTTCGGGAATCAGA′3, A16S1R 5′ TCACCGCAACATTCTGATTTG ′3) for amplification of a single DNA fragment of 356bp according to Wang et al. (2003).
DNA Extraction of Aeromonas hydrophila using QIA amp kit (Shah et al., 2009):
In Eppendorf tube, DNA template was prepared by boiling 200 μL of bacterial suspension in distilled water for 10 min. The tube was immediately cooled on ice and centrifuged (20000 RPM) at 4°C then the supernatant was kept at -20° until use.
DNA amplification reaction by using 16S rRNA for Aeromonas hydrophila (Stratev et al., 2016):
The amplification was performed on a Thermal Cycler (Master cycler, Eppendorf, Hamburg, Germany). PCR assays were adopted by using 25 μl of the reaction mixture contained 2X AmpliTaq DNA polymerase (Perkin-Elmer) – 12.5 μl, and 0.2 μl of 16S rRNA primers and 1 μl extracted DNA. Amplification conditions consisted of an initial denaturation at 95°C for 5 min, 50 cycles at 95°C for 30 sec, 59°C for 30 sec, 72°C for 30 sec, followed by final elongation at 72°C for 7 min. Following amplification, the amplified DNA fragments were analyzed by 2% of agarose gel electrophoresis (Applichem, Germany, GmbH) in 1X TAE buffer (0.04 M Tris, 0.02 M Acetic acid, 0.002 M Na2 EDTA) at 100 V for 45 min with 8 μl PCR product. Finally, the gel was stained with ethidium bromide and captured as well as visualized on UV transilluminator. A 100 bp plus DNA Ladder (Qiagen, Germany, GmbH) was used to determine the fragment sizes.
Experimental infection
Total Bacterial count
The drop plate technique for estimation of Aeromonas hydrophila strain per 1 ml was used in demonstration of the inoculum dose for the experimental studies as indicated by Herigstad et al. (2001).
Lethal Dose 50
A total number of 70 apparently healthy Oreochromis niloticus weighing (50±5 g) obtained from the hatchery and transported to the wet Lab. of Fish Diseases Department at Faculty of Fish and Fisheries Technology, Aswan University. The fish were divided into 7 groups, 10 fish/each group and the seventh group was kept as a control group. The fish were acclimated to laboratory conditions for two weeks under observation prior to injection for accommodation and to confirm that they are free from diseases. Fish were fed commercial diet twice a day at a feeding rate of 3% of its body weight.
Aeromonas hydrophila strain colony culture on TSA for 24 hours was utilized; the pure colonies were picked up and suspended in sterile saline in a tenfold serial dilution with incubation for 24 hours at 28 °C. Only the dilutions (102–107 cfu/mL) were used. The first six groups were intra-peritoneally injected with 0.5 ml/fish of each bacterial dilution. The fish in the seventh group were injected with 0.5 ml PBS/fish (Phosphate Buffer Saline). The fish groups were observed for one week post-inoculation. Clinical signs and mortalities were recorded twice/ day. The dead fish were subjected to postmortem examination to re-isolate the causative Aeromonas hydrophila strain from the internal organs. The LD50 (the dose which kill 50% of the injected fish) was calculated (Reed and Muench, 1938).
Pathogenicity test: A total number of 80 apparently healthy Oreochromis niloticus fish weighing (50±5 g), were divided into 4 groups; 20 fish per each. Each fish in the first group was intraperitoneally injected with 0.2 ml/fish of LD50 dose Aeromonas hydrophila strain which was determined previously (1.5x108). Each fish in the second group (control negative group), was intraperitoneally injected with 0.2 ml/fish of PBS (Phosphate Buffer Saline). Each fish in the third group (Control positive group), was intra-peritoneally injected with 0.2 ml/fish of got reference Aeromonas hydrophila strain (obtained kindly from the food analysis center, Faculty of Veterinary Medicine, Benha University, Egypt). The fish in the fourth group remained un-injected. Clinical signs and mortalities were recorded daily for 21 days post-inoculation. Moribund fish were bacteriologically examined to re-isolate the causative Aeromonas hydrophila strain from the internal organs and the freshly dead fish were moved for pm examination and histopathological studies. At the end of the experiment, all the survivors were sacrificed and examined as described above.
Histopathological Examination: Muscle, gills and liver tissue samples of experimentally infected fish with Aeromonas hydrophila were taken and fixed immediately in 10% neutral buffered formalin solution, dehydrated and blocked in paraffin then sectioned at 4 µm and stained with Hematoxilin & Eosin (H&E). The sections were examined under microscope using methods described by Bancroft and Gamble (2007).
Antimicrobial susceptibility assay of A. hydrophila (Antibiogramme):
Antimicrobial susceptibility for Aeromonas hydrophila strains was tested by the single diffusion method according to Stratev et al. (2015). Sensitivity discs with variable concentrations were used to determine the susceptibility of the isolated bacterial strains to different antimicrobial agents (Oxoid Limited, Basingstoke, Hampshire, UK) using Agar plate method on nutrient agar medium. The interpretations of the zones of inhibition were estimated as the maximal inhibition zone for the growth of microbe is said to that antibiotic had greatest impact on the microbe growth according to the guidelines stipulated by National Committee for Clinical Laboratory Standards “NCCLS” (2001). The tested strains were evaluated as susceptible, intermediate and resistant.
Multiple Antibiotic Resistance (MAR) index for each strain was determined according to Singh et al. (2010):
MAR index = No. of resistance (Isolates classified as intermediate were considered sensitive for MAR index) / Total No. of tested antibiotics
Results
Naturally infected Oreochromis niloticus revealed lethargy, anorexia, surface swimming, hemorrhages all over the body, detachment of scales, fin degeneration and erosions, distended abdomen and in some cases corneal opacity with exophthalmia (popeyes) (Figure 1A, 1B, Figure 2A, 2B, 2C). Meanwhile, postmortem examination revealed presence of sanguineous ascitic fluid in the abdominal cavity, gaseous intestinal contents, marbling (mosaic) appearance of gills with excessive mucus secretions and friable pale enlarged liver with distended gall bladder (Figure 2 D, 3A, 3B).
Phenotypic analysis of Aeromonas spp. colonies on TSA medium appeared round, convex, shiny and creamy while, on RS medium, appeared deep creamy or light yellow colonies with entire margin after 24 hrs of incubation. On Aeromonas base agar medium appeared small, dark green colonies with a dark center and on XLD medium appeared large, convex and round yellow colonies. On MacConkey’s agar medium appeared pale colonies then become pink while on blood agar medium it appeared large grayish circular, smooth, glistening colonies and surround by beta haemolysis. By Gram staining, Gram-negative cocco-bacilli to rod-shaped motile bacteria appeared. Aeromonas hydrophila is oxidase-positive, catalase-positive, Voges Proskauer-positive and lysine decarboxylase-positive bacteria while Aeromonas sobria is oxidase-positive, catalase-positive, Voges Proskauer-positive and lysine decarboxylase-negative bacteria.
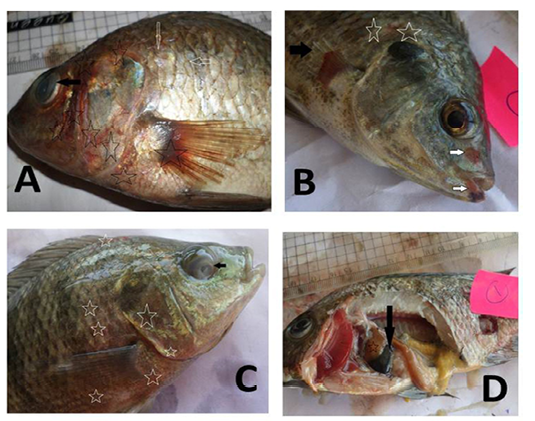
Figure 1: Oreochromis niloticus naturally infected with bacterial pathogens showing: A: Hemorrhages all over the body (stars) especially on anal fin (black arrow) and detachment of scales (white arrows). B: Caudal fin erosion (black arrow) and pectoral fin erosion (white arrow).
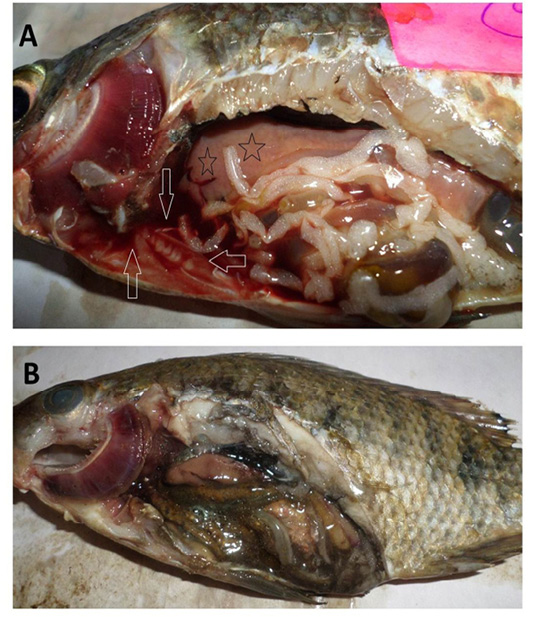
Figure 2: Oreochromis niloticus naturally infected with bacterial pathogens showing: A: Hemorrhages all over the body (stars), detachment of scales (white arrow) and cloudy eye (black arrow). B: Hemorrhages arround the mouth (white arrow), detachment of scales (stars) and erosion of pectoral fin (black arrow). C: Hemorrhages all over the body (stars) and cloudy eye (black arrow). D: Distended gall bladder (black arrow) and pale liver (star).
Pseudomonas sp. colonies on TSA medium appeared spindle shape and diffusible faint yellow-green fluorescence pigments produced after 24 hrs of incubation. On RS medium, appeared greenish in color after 24 hrs of incubation.
Table 1: Biochemical characters of different bacterial isolates obtained from naturally infected Oreochromis niloticus.
| Parameters | A. hydrophila | A. sobria | Ps. fluorescens | E. faecalis | Staph. aureus | Strept. agalactiae |
| Gram- stain | -ve | -ve | -ve | +ve | +ve | +ve |
| Motility test | Motile | Motile | Motile | Non motile | Non motile | Non motile |
| Oxidase | + | + | + | - | - | - |
| Catalase | + | + | + | - | + | - |
| Voges Proskauer | + | + | - | + | + | V |
| Urease | - | - | + | - | + | - |
| Ornithine Decarboxylation | + | - | - | - | + | - |
| Lysine decarboxylase | + | - | - | - | + | + |
| Arginine dihydrolase | + | - | + | - | - | + |
|
β galactosidase |
+ | + | - | - | - | - |
| Lactose fermentation | - | - | - | + | + | - |
| Glucose fermentation | + | + | + | + | + | + |
| Sucrose fermentation | + | + | + | + | + | + |
| Salicin fermentation | + | - | + | - | - |
- |
+ = Positive, - =Negative, v = Variable
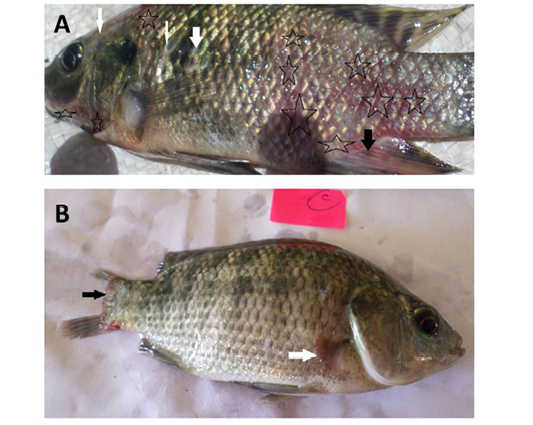
Figure 3: Oreochromis niloticus naturally infected with bacterial pathogens showing: A: Sanguineous ascitic fluid in the abdominal cavity (arrows), pale enlarged liver (stars) and marbling (mosaic) appearance of the gills. B: Gaseous intestinal content and marbling (mosaic) appearance of the gills with excessive mucus secretions.
On Pseudomonas-F agar, it appeared as 2-3 mm yellowish green colonies producing fluorescence after 48 hrs of incubation. By Gram staining, Gram-negative, rod-shaped motile bacteria motile recorded. Pseudomonas fluorescens is oxidase-positive, catalase-positive, and Voges Proskauer-negative bacteria.
Enterococcus sp. colonies on Streptococcus Selective agar showed very small rounded, white colonies with entire edges or creamy rounded, large colonies (2-3mm) have dew drops like. Enterococcus M broth medium color change was evaluated as an indicator of enterococci growth. An enterococci on BBLTM Enterococcosel Agar (EA) appeared dark brown and black colonies. By Gram’s staining, Enterococcus faecalis is Gram-positive arranged in pairs and sometimes short chains. Enterococcus faecalis is oxidase- negative, catalase-negative, and lysine decarboxylase-negative non motile bacteria.
Bacterial colonies of Staphylococcus aureus were golden yellow on TSA and MSA media with yellow halo surrounding the colonies on MSA. On TSI, yellow slant, yellow butt, no H2S and gas production were noticed. By Gram’s staining, Gram positive cocci accumulated in clusters appeared. Staphylococcus aureus is oxidase- negative, catalase-positive and lysine decarboxylase-positive non motile bacteria
Colonies of Streptococcus agalactiae on TSA agar appeared raised and glossy, with a diameter of 1.5-2.0 mm. On blood agar small white colonies, translucent or pigmented, slightly convex, up to 2 μm in diameter appeared. By Gram’s staining, Gram positive cocci non motile often in chains were noticed. Streptococcus agalactiae is oxidase-negative, catalase-negative and lysine decarboxylase-positive non motile bacteria. Other biochemical characteristics of these bacterial pathogens are given in Table 1.
From the complete bacteriological and biochemical examinations of the diseased Oreochromis niloticus, mainly six pathogenic bacteria, Aeromonas hydrophila, Aeromonas sobria, Pseudomonas fluorescens, Enterococcus faecalis, Staphylococcus. aureus and Streptococcus agalactiae have been associated with most of septicemic cases. Total prevalence of naturally infected Oreochromis niloticus with bacterial infections was (73%). The isolated bacteria were Aeromonas hydrophila (56%), Aeromonas sobria (45%), Pseudomonas fluorescens (33%), Enterococcus faecalis (11 %), Staphylococcus aureus (13%) and Streptococcus agalactiae (14%). Many cases of mixed infection in the same fish were detected. The most prevalent bacterial isolates were Aeromonas hydrophila (41 isolates). Randomly selected six isolates were checked by PCR. The outbreaks of the detected bacterial pathogens were usually during induced spawning in April and May each year.
Molecular characterization with conventional PCR to confirm biochemically identified Aeromonas hydrophila using 16S rRNA region, for the selected six Aeromonas isolates, resulted in an amplified product of 356 bp size (Figure 4).
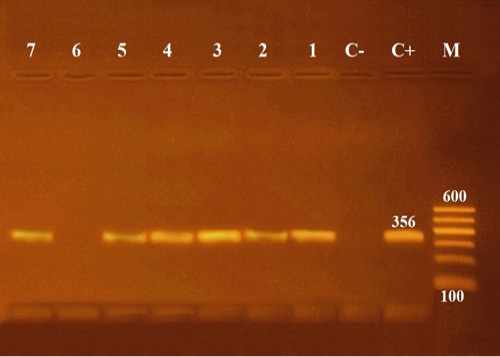
Figure 4: Agarose gel electrophoresis of PCR of 16S rRNA (356 bp) as species specific gene for confirmation of Aeromonas hydrophila isolated from Oreochromis niloticus. Lane M: 100 bp ladder as molecular size DNA marker. Lane C+: Control positive Aeromonas hydrophila for16S rRNA gene. Lane C-: Control negative. Lanes 1, 2, 3, 4, 5 & 7: Positive Aeromonas hydrophila strains. Lane 6: Negative Aeromonas hydrophila.
Experimental inoculation of Oreochromis niloticus with Aeromonas hydrophila isolated from naturally infected fish indicated that it is higly pathogenic and the cumulative mortality was 100 % in the challenged group within 7 days post inoculation. The fish began to die on the 2nd day post-challenge. The clinical signs and postmortem lesions of septicemic disease were observed in the experimentally infected Oreochromis niloticus with Aeromonas hydrophila which re-isolated from the internal organs of these fish. No clinical signs or mortalities were observed in the control group.
In the present study, the histopathological examinations of the experimentally infected Oreochromis niloticus with Aeromonas hydrophila, demonstrated interstitial edema in the muscular tissue, degeneration and loss of muscle architecture (Figure 5A). Meanwhile, the gills showed mononuclear cell infiltration in addition to epithelial lifting and loss of secondary lamellae (Figure 5B, 5C). In addition to, The hepatic tissue showed degeneration, loss of hepatocellular architecture, hepatocellular vacuolation and necrosis (Figure 5D, 5E, 5F).
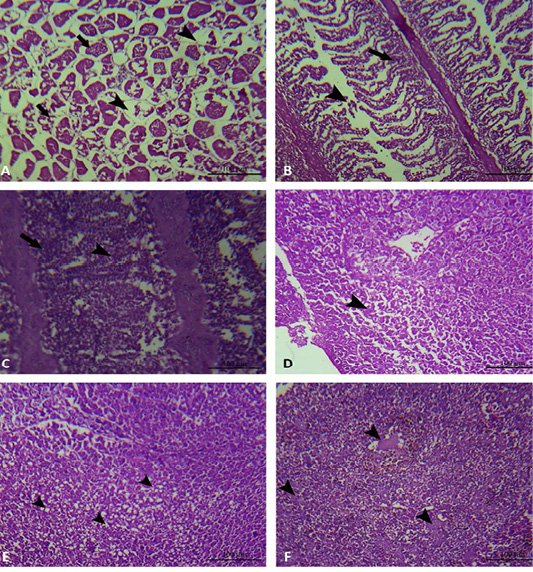
Figure 5: Muscle, gills and liver of experimentally infected Oreochromis niloticus with Aeromonas hydrophila. A: The muscular tissue showing interstitial edema (black arrow head), degeneration and loss of muscle architecture (black arrow). B and C: The gills showing mononuclear cell infiltration (black arrow), in addition to epithelial lifting and loss of secondary lamellae (black arrow head). D: The hepatic tissue showing degeneration and loss of hepatocellular architecture (black arrow head). E: The hepatic tissue showing hepatocellular vacuolation (black arrow head). F: The hepatic tissue showing hepatocellular necrosis (black arrow head).
Six isolates of Aeromonas hydrophila were screened against 13 antimicrobial agents to determine the susceptibility and the prevalence of multiple antibiotic resistance among these isolates (Table 2). The results indicated that the maj-
Table 2: Percentages of Antimicrobial susceptibility of Aeromonas hydrophila (n=6)
|
Antimicrobial agent
|
S (Sensitive) | I (Intermediate) | R (Resistant) | |||
| NO | % | NO | % | NO | % | |
| Penicillin (P) | - |
- |
- | - | 6 | 100 |
| Erythromycin (E) | - |
- |
- | - | 6 | 100 |
| Cloxacillin (CL) | - |
- |
1 | 16.67 | 5 | 83.33 |
| Oxytetracycline (T) | - | - | 2 | 33.33 | 4 | 66.67 |
| Ampicillin (AM) | 1 | 16.67 | 1 | 16.67 | 4 | 66.67 |
| Sulphamethoxazol (SXT) | 1 | 16.67 | 1 | 16.67 | 4 | 66.67 |
| Cefotaxim (CF) | 3 | 50 | - | - | 3 | 50 |
| Kanamycin (K) | 1 | 16.67 | 2 | 33.33 | 3 | 50 |
| Cephalothin (CN) | 2 | 33.33 | 1 | 16.67 | 3 | 50 |
| Nalidixic acid (NA) | 4 | 66.67 | - | - | 2 | 33.33 |
| Amikacin (AK) | 4 | 66.67 | - | - | 2 | 33.33 |
| Ciprofloxacin (CP) | 5 | 83.33 | - | - | 1 | 16.67 |
| Gentamicin (G) | 5 | 83.33 | - | - | 1 |
16.67 |
Table 3: Antimicrobial resistance profile of Aeromonas hydrophila strains (n=6).
| NO | Key No. | Antimicrobial resistance profile | MAR index |
| 1 | Liver 2 | P, E, CL, T, AM, SXT, CF, K, CN, NA, AK, CP, G | 1 |
| 2 | Spleen 4 | P, E, CL, T, AM, SXT, CF, K, CN, NA, AK | 0.846 |
| 3 | Kidney 10 | P, E, CL, T, AM, SXT, CF | 0.500 |
| 4 | Kidney 7 | P, E, CL, T, AM | 0.357 |
| 5 | Spleen 2 | P, E, CL, T, AM | 0.357 |
| 6 | Skin 1 | P, E | 0.143 |
|
Average 0.534 |
|||
P: Penicillin, E: Erythromycin, CL: Cloxacillin, T: Oxytetracycline, AM: Ampicillin, SXT: Sulphamethoxazol, CF: Cefotaxim, K: Kanamycin, CN: Cephalothin, NA: Nalidixic acid, AK: Amikacin, CP: Ciprofloxacin, G: Gentamicin.
ority of Aeromonas hydrophila isolates is sensitive to Gentamicin, Ciprofloxacin, Amikacin, Nalidixic acid, Cefotaxim, Cephalothin, Kanamycin, Sulphamethoxazol and Ampicillin (in descending order). However, two strains are moderately sensitive to Oxytetracycline and one strain is moderately sensitive to Cloxacillin. All six isolates are completely resistant to Penicillin and Erythromycin (Table 2). In our study, the tested isolates showed resistant patterns for many antibiotics recording MAR index of 0.534 (Table 3).
Discussion
Protecting the aquaculture industry from bacterial diseases is a priority because of the high economic losses caused by their outbreaks. The accuracy of disease diagnosis in fish results in success of disease control, prevention and treatment.
The infected fish showed lethargy and sluggish movement and this could be a result of anorexia as well as frayed and sloughed fins which subsequently affected the vital activities and swimming of the infected fish. The hemorrhages all over the body may be due to the hemolysin secreted by the isolated bacteria which make hemolysis of the RBCs as well as, elastase enzyme which contribute significantly to vascular damage because the blood vessels are mainly composed of elastic and collagenous fibers. The clinical signs displayed in the current study were nearly corresponded with that recorded by previous authors (Ahmed and Shoreit, 2001; El-Ashram, 2002; Khafagy et al., 2009; Yardimci and Aydin, 2011; Noor El-Deen et al., 2014; Soliman et al., 2014; Othman et al., 2015; Dar et al., 2016; Abd El-Tawab et al., 2017; El-Barbary, 2017; Laith et al., 2017; Osman et al., 2017; Pech et al., 2017; Saleh et al., 2017; Amrullah et al., 2018; Khalil and Emeash, 2018).
The post mortem changes may be attributed to the isolated septicemic bacteria and their virulence genes. Pathogenic bacteria might secrete extracellular products as aerolysin, haemolysin, cytotoxic toxins, S-layers and extracellular enzymes as gelatinase, proteases, lipases, hyaluronidase, nuclease hypuricase, elastase, C5a-ase, CAMP factors and aggregation substance. These secretions possess cytolytic, enterotoxic and haemolytic functions responsible for pathogenesis of these bacteria (Pritchard and Lin, 1993; Khafagy et al., 2009; Umesha et al., 2011; Soliman et al., 2014; Dahdouh et al., 2016; Osman et al., 2017; Ahmed et al., 2018; Mansour et al., 2019). The postmortem findings displayed in this study are nearly similar to those reported by many previous authors (Ahmed and Shoreit, 2001; Musa et al., 2009; Pretto-Giordano et al., 2010; Abd El-Tawab et al., 2017; Saleh et al., 2017; Ortega et al., 2017).
The colonies morphology, Gram staining and the biochemical profile of the detected bacteria are nearly identical to those reported by many other previous authors ( Ahmed and Shoreit, 2001; Masbouba, 2004; Khafagy et al., 2009; Noga, 2010; Soliman et al., 2014; Soliman et al., 2016; Dar et al., 2016; Abd El-Tawab et al. 2017; Abd El-Kader and Mousa-Balabel, 2017; Osman et al., 2017; El-Barbary, 2017; Laith et al., 2017; Ortega et al., 2017; El-Gamal et al., 2018; Hardi et al., 2018).
The prevalence of naturally infected Oreochromis niloticus with bacterial infections is (73%) and the most prevalent bacteria is Aeromonas hydrophila. These findings may differ completely or partially with that recorded by many authors and this could be due to abiotic and biotic conditions of the environments where the studies performed.
Most of bacterial pathogens in fish farms are normal inhabitant in water and do not develop simply as the result of exposing a host to an infectious agent (Wedekind et al., 2010). The outbreaks of the detected bacterial pathogens were usually associated with change in environmental conditions (stressors) as over wintering of the Oreochromis niloticus, sudden change in temperature, inappropriate handling and physical injury during transportation and induced spawning in April and May each year. Environmental stressors can affect the homeostatic mechanism of fish, thus reducing their resistance to pathogenic organisms and triggering the disease outbreaks (Small and Bilodeau, 2005) so inapt farm management systems are prone fish in aquaculture to a variety of diseases (El-Sayed, 2006). Some bacterial fish diseases are uncommon to infect tilapia as Methicillin-resistant Staphylococcus aureus and Aeromonas sobria but due to close contact of aquaculture environment with animal and/or human wastes, these bacteria flourish (Soliman et al., 2014; Dar et al., 2016).
Polymerase chain reaction has been proven to be more accurate and rapid method for identification of bacterial pathogens. The 16S ribosomal RNA gene is a highly conserved region present in bacteria which plays a major role in gene coding. It is considered as a standard marker for bacterial phylogenetic analysis to differentiate the species (Nagpal et al., 1998). The molecular characterization of Aeromonas hydrophila using16S rRNA region agreed with that described by Stratev et al. (2016).
Experimental infections of Oreochromis niloticus with Aeromonas hydrophila revealed that it is highly pathogenic to this fish species and the obtained result agreed with that recorded by Zhang et al. (2016).
The pronounced destructive changes in the musculature of experimentally infected Oreochromis niloticus with Aeromonas hydrophila may be due to protease activity of the isolated bacteria, which possess also ability to adhere to cells (Azad et al., 2001; Soto, 2009) and the histopathological changes in the gills and liver were similar to previous records by many authors (Azad et al., 2001; El-Ashram, 2002; Soto, 2009; Yardimci and Aydin, 2011; Noor El-Deen et al., 2014; Abdelhamed et al., 2017; El-Barbary, 2017; Al-Yahya et al., 2018) in Oreochromis niloticus and other fish species. The pathogenicity of Aeromonas hydrophila for experimentally infected Oreochromis niloticus may be attributed to the production of lethal toxins and extracellular products such as hemolysin, protease and elastase produced by this bacteria (Afifi et al., 2000; Yardimci and Aydin, 2011; Noor El Deen et al., 2014; El-Barbary, 2017). The histological changes due to Aeromonas hydrophila become distinct only if clinical conditions are prolonged (Paperna, 1996).
Studies of the of bacterial pathogens sensitivity to antibiotics in fish are of major time-wise and important for the development of new chemotherapeutic agents to combat bacterial infections in certain cultured fish population. Our results are nearly similar to those reported by Dahdouh et al. (2016) who declared that Aeromonas hydrophila isolated from freshwater fish was sensitive to Enrofloxacin, Ofloxacin, Gentamicin and resistant to Erythromycin. El-Barbary (2017) mentioned that Aeromonas hydrophila showed susceptibility to Ciprofloxacin, Norfloxacin, Gatifloxacin, Lomefloxacin, Gentamycin, Kanamycin and resistance to Penicillin and Erythromycin. On the other hand, Ahmed and Shoreit (2001) found that Aeromonas sp. isolated from Oreochromis niloticus from Aswan fish hatcheries were high sensitive to Oxytetracycline which showed resistance to most isolates of Aeromonas hydrophila in our study and this indicate the development of drug resistance to Oxytetracycline in this area due to the indiscriminate use of antimicrobials in aquaculture.
MAR index of Aeromonas hydrophila is 0.534 and has been indicative for the wide antibiotics usage and this agree with that reported by Laith and Najiah (2013) who declared that the MAR index for Aeromonas hydrophila isolates was ranged from 0.10-0.50 as well as, Orozova et al. (2010) who declared that when the MAR index ≥0.2, this indicate the wide antibiotics usage. Ahmed et al. (2018) mentioned that MAR index for Aeromonas hydrophila isolates was 0.489. Perretta et al. (2018) recorded that the isolates of Aeromonas hydrophila that had MAR rates higher than 0.2 came from fish farms that routinely use antimicrobials. Increasing cases of multi-resistance among Aeromonas sp. isolates may be due to the horizontal transfer of effectively genetic elements like plasmids and class 1 integrones for antibiotic resistance from and to other enterobacteria and furthermore by the indiscriminate use of antimicrobials in aquaculture (Marchandin et al., 2003; Jacobs and Chenia, 2006). Aeromonas hydrophila showing multiple antibiotic resistance (MAR) is a worldwide problem caused by the misuse of antibiotics and consider a public health hazard (Kaskhedikar and Chhabra, 2010; Odeyemi et al., 2012; Sharma et al., 2015; Ahmed et al., 2018; Perretta et al., 2018).
In conclusion, Aeromonas hydrophila, Aeromonas sobria, Pseudomonas fluorescens, Enterococcus faecalis, Staphylococcus aureus and Streptococcus agalactiae are the most significant microbial agents affecting Oreochromis niloticus in Sahary fish hatchery, Aswan governorate (southern Egypt). Stressors adversely affect Oreochromis niloticus and make them more susceptible to different bacterial diseases so best management practices must be adopted. The development of multi antibiotic resistance among clinical isolates of Aeromonas hydrophila is a potential risk for the fishery farms as well as, underlines the necessity of a constant monitoring of their resistance range. Aeromonas hydrophila is the most prevalent bacterial species, causing septicemia and mortalities that lead to serious economic losses. Aeromonas hydrophila is sensitive to Gentamicin, Ciprofloxacin, Amikacin and Nalidixic acid.
Conflict of Interest
The authors declare no conflicts of interest.
authors contribution
All authors contributed equally.
References






