Comparative Analysis of Muscle Development in Zebrafish with Different Intermuscular-Bo nes Patterns
Comparative Analysis of Muscle Development in Zebrafish with Different Intermuscular-Bo
nes Patterns
Jian Yang1, Guangxiang Tong2, Zhipeng Sun2, Xianhu Zheng2, Weihua Lv2, Dingchen Cao2, Xiaowen Sun2 and Youyi Kuang2*
1National Demonstration Center for Experimental Fisheries Science Education, Shanghai Ocean University, Shanghai 201306, China
2Heilongjiang River Fisheries Research Institute, Chinese Academy of Fishery Science, Harbin, 150070, China
ABSTRACT
In this study, we used an intermuscular-bones (imbs) partial deletion zebrafish (Danio rerio) mutant to analyze the impact of imbs deficiency on adjacent muscle development and growth by examining expression of muscle-specific genes and muscle structure. Five muscle-specific genes including myod, myog, myf5, mef2ca and sox6 were selected to test and verify the expression differences in embryonic development stages (3 hpf, 6 hpf, 12 hpf, 24 hpf, and 72 hpf) and post-embryonic stages (15 dpf, 30 dpf, 45 dpf, 60 dpf and 75 dpf). Compared to the wild-type (WT) siblings, the mutants showed no significant differences in the 5 gene expressions. Among different development stages, the expression levels and patterns of the 5 genes in the mutants were similar to that of WT zebrafish in both embryonic and post-embryonic development stages. Furthermore, the results of histological analyses of the muscle fiber showed that there were no significant differences in muscle fiber density between imbs mutant and WT zebrafish, and no significant differences between anterior and posterior part to the dorsal fin at the same developmental stages. In WT and mutants, the density of muscle fiber declined gradually over time. In conclusion, the lack of intermuscular-bones has no influence on adjacent muscle development.
Article Information
Received 25 November 2018
Revised 22 June 2019
Accepted 17 September 2019
Available online 24 December 2020
Authors’ Contribution
YK and XS conceived the study. GT carried out the genetic screen of mutant. YJ and YK wrote the manuscript. JY, ZS and DC carried the histological analysis. JY, XZ and WL carried out the qRT-PCR analysis.
Key words
Zebrafish, Intermuscular-bones, Muscle development, Muscle-specific genes, Muscle fiber
DOI: https://dx.doi.org/10.17582/journal.pjz/20181125111102
* Corresponding author: [email protected]
0030-9923/2021/0001-0313 $ 9.00/0
Copyright 2021 Zoological Society of Pakistan
INTRODUCTION
Intermuscular-bones (imbs) are small bones in muscles on the both sides of the vertebra, which are ossified from myoseptum (Patterson et al., 1995; Meng, 1987), which are popular in freshwater fish species (Lv et al., 2007), such as common carp and the four other major Chinese carps (Mylopharyngodon piceus, Ctenopharyngodon idellus, Hypophthalmichthys molitrix, Hypophthalmichthys nobilis). These bony structures are inconvenience for producing flesh products, as well as reducing the flesh quality because of the difficulty to remove. At present, most of the studies on imbs focus on using them as features for classification (Rom et al., 1975; Johmn et al., 2001), distribution (Dong et al., 2006; Li et al., 1987, 2013; Bing, 1962; Gao, 1984) and morphological development (Ke et al., 2008; Karsenty et al., 2002: Lv et al., 2012). Molecular and genetic analyses of the imbs’ development are still rare (Wan et al., 2016; Nie et al., 2017; Liu et al., 2017), especially on their specific function as a tissue.
Studies on the histology and calcification of imbs in fishes with different swimming modes (e.g. zebrafish, and Japanese eel, Anguilla japonica) have shown that imbs act as structural support to the muscles and transmission of strength (Yao et al., 2015). However, fish with the same swimming mode as zebrafish, crucian carp (Carassius cuvieri), blunt snout bream (Megalobrama amblycephala), bighead carp (Aristichthys nobilis) and silver carp (Hypophthalmichthys molitrix), have significant differences in number and form. Crucian carp has less imbs than the other three fishes and might rely more on muscle than imbs to swim compared to blunt snout bream, bighead carp and silver carp (Dong et al., 2006; Li et al., 2013; Li et al., 2017). Studies of zebrafish have also shown that strength transmission rely mainly on muscle fiber and not imbs (Sun, 2008). Therefore, we speculate that imbs might not be necessary for fish swimming. However, there is no study to verify the developmental relationship between muscle fiber and imbs. As suggested by lack of imbs in teleost species such as catfish (Silurus asotus Linnaeus) and tilapia (Oreochromis niloticus), inhibition of imbs formation probably will not impact the survival. It is unconfirmed that imbs’ deficiency would be harmful to muscle attachment.
Zebrafish muscle contains two kinds of muscle fiber: one is slow twitch muscle fibers which are needed for slow-swimming; the other is fast twitch muscle fiber required for fast swimming (Stickney et al., 2000). Both types of muscles develop immediately from progenitor cells of mesoblast at early stage of embryonic development. Myogenic regulatory factor (MRF) family genes include myod, myf5 and myog, which regulate and control the specification and differentiation of muscle cells (Pownall et al., 2002). Transcription factors, myod and myf5, are markers of muscle precursor cells (Coutelle et al., 2001; Weinberg et al., 1996), both of them are expressed in developing somites, and are essential for initiating the skeletal muscle program in an embryo. Gene myf5 initiates skeletal muscle development at gastrula stage, whereas myog expresses later than myf5 and functions in both myoblast and skeletal muscles. Gene myod plays an important role in differentiation and maturation of muscle fibers (Watabe, 1999; Tan et al., 2002; Iban et al., 2012: Daniel et al., 2008). Gene mef2 is an important regulator in skeletal muscle differentiation (Ticho et al., 1996: Olson, 1992). In all known skeletal muscle development processes, myf5/myod expression is followed by upregulation of myog and mef2 family factors; the latter’s enhancing expression of differentiation genes (Yun and Wold, 1996). Gene sox6 is a key transcriptional regulator of fast-twitch muscle fiber differentiation in the zebrafish and ectopic over-expression of sox6 is sufficient to downregulate slow-twitch specific gene expression in zebrafish embryos (Harriet et al., 2015). These 5 genes are muscle-specific expression genes and could be used to indicate the development of muscle.
With the advantage of forward and reverse genetic technologies, it becomes feasible to study the relationship between imbs and muscles during early development. Through genetic screens, we obtained a mutant with partial loss of imbs, which is homozygous and fertile (Fig. 1). Using this imbs mutant, we analyzed the impact of abnormality or loss of imbs to the development of muscles in two aspects, the expression of 5 muscle-specific genes, myod, myf5, myog, mef2ca and sox6, and the histological differences of muscle fibers.
MATERIALS AND METHODS
Lineage of experimental zebrafish
In the previous studies, we screened an imbs partial deletion mutant lineage and established homozygous lines which were demonstrated through classical Mendelian inheritant experiments that the mutants were homozygous recessive. To eliminate the impact resulted from different genetic backgrounds, we established the experimental lineages using a strategy illustrated in Figure 2 and described as follows. First, we crossed the mutant and wildtype zebrafish to construct F2 generation families. We used bone staining method to check the phenotypes of F2 families and found that the mutational phenotypes occupied about 25% individuals which are consistent with Mendel’s law; it confirmed the results in the previous studies that the imbs mutants were homozygous recessive. Among F2 families, healthy individuals were inter-crossed to establish F4 families, after characterizing phenotypes of F4 families by bone-staining, 23 homozygous imbs mutant families and 13 homozygous wildtype families were chosen to carry out the studies.
Fish culture and breeding
Families of wild-type and mutant were chosen as female-male ratio 1:1 to conduct the breeding experiments. After spawn, eggs in each family were counted and hatched in one culture dish at 28℃. The dead eggs or abnormal larvae were counted and clean up each day. The breeding experiments were replicated four times in four weeks using the same parents. The fertilized rate, hatched rate and deformed rate were represented with the mean value of the four replicates.
Growth measure
After 7dpf, larvae of WT and imbs mutant were divided randomly into three parallel groups, each containing 30 individuals. Fish in each group were cultured in polystyrene plastic tanks (27.5×23.5×19 cm) containing 8L water at water temperature 28℃ and fed with fairy shrimp purchased from Tianjin Fengnian Aquaculture LTD. Growth of individuals in different imbs phenotype group was measured at 5 development stages (15 dpf, 30 dpf, 45 dpf, 60 dpf and 75 dpf). For biometric analysis, fish were anesthetized with tricaine (0.1mg/mL), weighed, and measured in lateral decubitus for standard length measurements.
Gene expression
Gene expressions were tested in 5 embryonic development stages (3 hpf, blastula; 6 hpf, midgastrula; 12 hpf, segmentation period; 24 hpf, pharyngula; and 72 hpf, hatching period) and 5 post-embryonic development stages (15 dpf, 30 dpf, 45 dpf, 60 dpf and 75 dpf). In embryonic development, we sampled wild-type and mutant fertilized eggs or larvae, each sample in the same stage were triplicate; each sample contained 30 fertilized eggs in 3 hpf, 6 hpf, 12 hpf and 24 hpf, and 10 larvae in 72 hpf. In post-embryonic development, we dissected muscles of anterior and posterior to dorsal fin separately in 5 stages. After anesthetization, fish was placed on ice for muscle sample collection. The samples were collected in triplicate for each period.Total RNA was extracted from each sample using Trizol reagent (Invitrogen, CA, USA), and cDNA was synthesized using High Capacity cDNA Reverse Transcription kits (Roache, CA, USA).
Five muscle-specific genes including myod, myog, myf5, mef2ca and sox6 were chosen to assess their expression by qRT-PCR. The primers for amplification were designed using Primer3 program and are listed in Table I. Quantitative real time PCRs were performed using Luna Universal qPCR Master Mix M3003 kits (NEB, MA, USA) with10 µL reaction volume which contained 5 µL 2×qPCR mix, 0.25 µL 10 µM forward and reverse primers, 1µL 50ng/µL cDNA template, 3.5 µL nuclease-free water. The amplification program was set up as follows: 95℃ pre-denaturation 60 s, followed by 40 cycles each of 95℃ denaturation 15 s, 60℃ extension 30 s. Expression of target genes was normalized against reference gene gapdh.
Table I. Muscle-specific gene primers for qRT-PCR.
|
Gene bank |
Gene |
Sequence |
|
NM_001328013.1 |
myod |
F: 5'TCCGAGGACATGAGCCAGAT3' |
|
R: 5'GACGCCGTTTTGCCTGAATA3' |
||
|
NM_131006.1 |
myog |
F: 5'AGAGACCTCAGGTTGGATTGC3' |
|
R: 5'TCCTCTAGTGATCAGGGCTCT3' |
||
|
NM_131576.1 |
myf5 |
F: 5'GCGTCAAAGTTGTAGCTATTCCC3' |
|
R: 5'TACTACAGCCTGCCGATGGA3' |
||
|
NM_131301.2 |
mef2ca |
F: 5'CTCTTTCCGTCTGTGCCTCT3' |
|
R: 5'CCGAGGAAGAGAAAGCACCA3' |
||
|
NM_001123009.1 |
sox6 |
F: 5'TCGTGTGGAAAAATGGGGATCA3' |
|
R: 5'ATCGCCAGACAACAGCAGCA3' |
||
|
NM_001115114.1 |
gapdh |
F: 5'ACCCGTGCTGCTTTCTTGAC3' |
|
R: 5'GACCAGTTTGCCGCCTTCT3' |
Muscle histology
Muscles of anterior and posterior to dorsal fin at 5 stages, 15 dpf, 30 dpf, 45 dpf, 60 dpf and 75 dpf were used for histological analysis. Muscle tissues were fixed in Bouin’s solution for 4h and preserved in 70% ethanol. Histological samples were prepared using classic methods (Li, 2009). The samples were cut into 6 μm sections, stained with hematoxylin/eosin, and analyzed for density and diameter of muscle fibers.
A standard circle was used for counting the number of muscle fibers and calculating muscle fibers’ density. Samples at 15 and 30 dpf were measured using a standard circle of radius 30.0 μm (area 2827.4 μm2) considering the small size of the sample. For samples at 45, 60 and 75 dpf a standard circle of radius 50.0 μm (area 7853.9 μm2) was used in the measurement. Density of muscle fibers was calculated using the formula as follows:
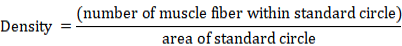
Statistical analyses
Differences of growth, gene expression, muscle fiber density and diameter among 5 growth stages were used one-way ANOVA to analyze the differences among stages, and differences between mutant and WT were analyzed with two-way ANOVA followed by Bonferroni posttest. Relative gene expression was calculated using the 2-△△CT method (Kenneth et al., 2001).
RESULTS
Embryonic development
Fertilization rate, hatching rate and deforming rate of wild-type zebrafish were 0.908±0.095, 0.902±0.072 and 0.024±0.011, respectively. Rate of fertilization, hatch and deformity of mutant individuals were 0.927±0.051, 0.933±0.061 and 0.021±0.018, respectively. The analysis revealed that there were no significant differences between wild-type and mutant zebrafish in embryonic development (Fig. 3).
Growth
The weight and body length of the experimental fish increased dramatically along with their growth. The two-way ANOVA analysis showed that there was no significant difference between WT and mutants in body weight and body length at of 5 developmental stages measured. The survival of the animals was 100% during the growth experiment (Fig. 4).
Gene expression in embryonic development
Expression of 5 muscle-specific genes in 5 embryonic developmental stages increased at the beginning, and then was reduced later on. The expression of myf5 and myod reached a peak at 12hpf, and the other 4 genes reached the peak at 24 hpf. Furthermore, mef2ca, myog and sox6, had a low expression level in blastula and gastrula period, and reached a peak in segmentation and pharyngula period. Comparison between wild-type and mutant samples showed no significant differences of the 5 gene expressions in the same developmental stages (Fig. 5).
Gene expression in post-embryonic development
In general, there are no significant differences in the expression of 5 muscle-specific genes between imbs mutant and WT in the same anatomical locations of individual at the same developmental stages. The expression level of each gene in anterior and posterior to the dorsal fin did not seem much different. Moreover, the expression level of each gene at the same anatomical location decreased gradually as development proceeded (Fig. 6).
Muscle histology
The muscle histological analyses showed that there was no significant difference in the density of muscle fibers between imbs mutant and WT in the same anatomical locations at the same growth stages. The number of muscle fibers in a unit area at the anterior area to dorsal fin was less than the posterior one. Besides, the density of muscle fibers at the same location decreased gradually along with the growth of individuals.
The diameters of muscle fibers showed the same trend as the density of muscle fibers. There were also no significant differences between WT and imbs mutants in the same anatomical locations at the same developmental stages (Fig. 7, Table II).
Table II. Diameter of muscle fibers at different locations of the zebrafish.
|
Growth stage |
Group |
Diameter of anterior dorsal fin (μm) |
Diameter of posterior dorsal fin (μm) |
|
15 dpf |
WT |
12.13 ± 1.76a |
11.47 ± 0.90a |
|
Mutant |
13.07 ± 1.90a |
11.93 ± 0.35a |
|
|
30 dpf |
WT |
15.87 ± 0.58b |
12.70 ± 0.75b |
|
Mutant |
16.33 ± 0.91b |
13.27 ± 0.60b |
|
|
45 dpf |
WT |
17.13 ± 0.91c |
15.03 ± 0.45c |
|
Mutant |
17.03 ± 0.57c |
14.90 ± 0.79c |
|
|
60 dpf |
WT |
17.83 ± 1.45d |
16.27 ± 0.81d |
|
Mutant |
18.33 ± 1.79d |
16.10 ± 0.53d |
|
|
75 dpf |
WT |
19.40 ± 1.18e |
17.37 ± 1.12e |
|
Mutant |
19.27 ± 1.38e |
17.07 ± 1.03e |
Data on different body location were analyzed separately. Diameter of muscle fibers are expressed as mean ± SD. Significant differences (P<0.05) occurred between different development stages represented by different letters (a, b, c, d and e), while there were no significant difference between imbs mutants and WT individuals represented by the same letter.
DISCUSSION
Development of somites and muscles in fish is similar to that of amphibians, birds and mammals (Kimmel et al., 1995). Precursors of adult slow- and fast-twitch muscle fibers already emerge in early embryo development. By the end of segmentation period, fast-twitch muscle fibers move into deeper areas of myotomes, and slow-twitch muscle fibers form a monolayer on the surface of myotome.
Slow- and fast-twitch muscle fibers further differentiate to become red muscle and white muscle, respectively in the end (Sun, 2008). MRFs (myod, myf5, myog) and mef2ca play a great role in differentiation and maturation of skeletal muscle cells. Therefore, myod and myf5, expressed in developing somites, are essential for initiating the skeletal muscle program in the embryo (Coutelle et al., 2001; Weinberg et al., 1996), whereas myod/myf5 expression is followed by an up-regulation of myog and mef2ca family factors (Yun and Wold, 1996). As for sox6, studies on zebrafish and mice show that it displays an increase in slow-specific gene expression and a concomitant decrease in the expression of fast-twitch specific genes suggesting that sox6 normally functions to promote the fast-twitch differentiation and repress slow-specific gene expression in fetal muscle fibers (Hagiwara et al., 2007; An et al., 2011; Quiat et al., 2011; Von Hofsten et al., 2008: Harriet et al., 2015). Gene expression indicated that all five genes were increased in first and decreased the last during five periods of embryonic development. The expression of myf5 and myod reached a peak in 12hpf (segmentation period), and maintained a high level in 24 hpf (pharyngula period). Besides, gene mef2ca, myog and sox6 have a low expression level in blastula and gastrula period, and reached a peak in segmentation and pharyngula period. The results agreed well with the previous literature. Moreover, most studies focus on the expression of MRFs during embryonic development period. Few studies discussed about the expression in juvenile and adult fish. In our study, we explore the expression of 5 muscle-specific genes in post-embryonic development. The results showed that the expression of 5 genes decreased along with the growth of zebrafish. As the growth of fish and the maturation of muscle fibers, the expression of genes functioning in development of muscle declined slowly which might result in increase in the density and diameter of muscle fiber slowly in the adult zebrafish individuals.
As for the difference of muscle development between anterior to posterior to dorsal fin, this study showed that the expression level of five muscle-specific genes was close to each other which indicated that the development pattern of muscle in anterior to dorsal fin was similar to that of muscle of posterior to dorsal fin. But the density of muscle fibers of posterior to dorsal fin was a little larger than anterior ones, and the diameter of posterior ones was smaller than anterior ones, which might be because of the tail part of fish which required more muscle fibers to provide strength to fish movement compared to front part. The development pattern of muscle is still influenced by its functional requirement.
With regard to the influence of imbs elimination to the development of muscle fiber, our study showed that no negative affect occurred on muscle development from three aspects. In gene expression aspect, there are no significant differences between imbs mutant and WT at the same part of zebrafish during the same growth stage. In muscle histology aspects, there is no significant difference in the muscle fibers’ density between imbs mutant and WT of the same part and growth stage. In the aspect of embryonic development and post-embryonic development growth, there was no significant difference between WT and mutants in the rate of fertility, hatching and deformities, body weight and body length. Therefore, we speculated that the elimination of imbs did not affect muscle-specific genes’ expression and may not have detrimental influence on muscle’s development.
Researchers began to explore the value of eliminating imbs in fish as early as the 1960s. Available studies show that significant difference occurs among fishes of different species and ploidy. Some researches have shown that the existence of imbs is closely related to the evolution of fish. The number of imbs became larger as the teleost appeared. Along with the evolution of fish, the number of imbs decreased gradually; even disappeared completely in some fishes (Patterson et al., 1995; Ma et al., 2012). Moreover, studies show that because of small body and swim bladder, zebrafish have no need of strong bones but powerful muscles to support body movement. Therefore, primary somites of zebrafish are myotomes, not sclerotomes (Sun, 2008). Some researchers also speculate that imbs might be kind of rudimentary tissues (Lv et al., 2007). Therefore, imbs were considered that they only have limited supplementary role in supporting muscles and transmitting strengthen, and the elimination of imbs might not have negative effect on body shape, tissue structure and life activity. However, there are no sufficient number of examples to draw a definitive conclusion that there are no disadvantages after the deletion of imbs in fish with imbs. Through genetic screen methods, we could establish the imbs deletion model to investigate the role of imbs. In this study, we demonstrated that the lack of imbs didn’t impact the muscle development and growth of zebrafish, while it is still unknown whether imbs’ deletion would have an influence on reproductive performance, avoiding predators, predation and other living activities.
CONCLUSION
In conclusion, intermuscular-bone is a rudimentary organ according to speciation evolution study. Our results indicated that the deletion of Imbs had no influence on the embryonic development and growth and there was no significant difference in muscle structure and development no matter in embryonic or post-embryonic stages. However, it is still unconfirmed whether the deficiency of imbs would be pernicious to fish which already have imbs, and more studies should be carried out to access the impact of the deficiency of Imbs.
ACKNOWLEDGEMENTS
The authors would thank Dr. Yi Zhou from Boston Children’s Hospital of Harvard Medical School for providing useful comments and revising the manuscript. This study is financially supported by the National Key R&D Program of China(2018YFD0900102); Central Public-interest Scientific Institution Basal Research Fund, HRFRI(HSY201802Z); Central Public-interest Scientific Institution Basal Research Fund CAFS(no. 2019XT0102).
Statement of conflict of interest
The authors declare there is no conflict of interest.
REFERENCES
An, C.I., Dong, Y. and Hagiwara, N., 2011. The intermuscular bones and ligaments of teleostean fishes. BMC Dev. Biol., 11: 59.
Bing, Z., 1962. On the myoseptal spines of the carp (Cyprinus carpiol). Curr. Zool., 14: 175-178.
Coutelle, O., Blagden, C.S., Hampson, R., Halai, C., Rigby, P.W.J. and Hughes, S.M., 2001. Hedgehog signaling is required for maintenance of myf5 and myod expression and timely terminal differentiation in zebrafish adaxial myogenesis. Dev. Biol., 236: 136-150. https://doi.org/10.1006/dbio.2001.0193
Daniel, J., Macqueen, Ian, A. and Johnston, 2008. An update on myod evolution in teleosts and a proposed consensus nomenclature to accommodate the tetraploidization of different vertebrate genomes. PLoS One, 2: 1-9. https://doi.org/10.1371/journal.pone.0001567
Dong, Z.J., Huang, D.Z., Li, L.J., Yuan, X.H., Liao, W.M., Chen, Q.Q., Lu, Z.B. and Zhang, B.L., 2006. Preliminary study on intermuscular bones of several cultured cypriIlids. J. Shanghai Fish. Univ., 15: 425-429.
Gao, W.J., 1984. A study on the morphological and distribution of os intermuscular of Ctenopharyngodon idellus. J. Hebei Agric. Univ., 7: 178-184.
Hagiwara, N., Yeh, M. and Liu, A., 2007. Sox6 is required for normal fiber type differentiation of fetal skeletal muscle in mice. Devel. Dyn., 236: 2062-2076. https://doi.org/10.1002/dvdy.21223
Harriet, E., Jackson, Yosuke, O., Wang, X.G., Stone, E., Vincent, T.C. and Philip, W.I., 2015. The role of sox6 in zebrafish muscle fiber type specification. Skelet. Muscle, 5: 1-17. https://doi.org/10.1186/s13395-014-0026-2
Iban, S., Nathalie, S. and Jean C.G., 2012. Myostatin inhibits proliferation but not differentiation of trout myoblasts. Moll. Cel. Endocrinol., 351: 220-226. https://doi.org/10.1016/j.mce.2011.12.011
Johnson, G.D. and Patterson, C., 2001. The intermuscular system of acanthomorph fishes: A commentary. Am. Mus. natl. Hist., 3312: l-24. https://doi.org/10.1206/0003-0082(2001)312<0001:TISOAF>2.0.CO;2
Karsenty, G. and Wagner, E.F., 2002. Reaching a genetic and molecular understanding of skeletal development. Dev. Cell, 2: 389-406. https://doi.org/10.1016/S1534-5807(02)00157-0
Kenneth, J., Livak, Thomas, D. and Schmittgen, 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-△△CT method. Methods, 25: 402-408. https://doi.org/10.1006/meth.2001.1262
Ke, Z.H., Zhang, W., Jiang, Y. and Bao, B.L., 2008. Developmental morphology of the intermuscular bone in Hypophthalmichthys molitrix. J. Chin. Zool., 43: 88-96.
Kimmel, C.B., Ballard, W.W., Kimmel, S.R., Ballard, Seth, R.K., Bonnie, U. and Thomas, F.S., 1995. Stages of embryonic development of the zebrafish. Dev. Dynam., 203: 253-310. https://doi.org/10.1002/aja.1002030302
Li, A.H., 2009. A modified hemetoxylin and eosin staining method. Chin. Mod. Med., 10: 53-54, 73.
Li, L., Zhong, Z.Z., Zeng, M., Liu, S.J., Zhou, Y., Xiao, J., Wang, J. and Liu, Y., 2013. Comparative analysis of intermuscular bone in different ploidy fish. Sci. China Life Sci., 3: 189-200.
Liu, H., Chen, C.H., Gao, Z.X., Min, J.M., Gu, Y.M., Jian, J.B., Jiang, X.W., Cai, H.M., Ebersberger, I., Xu, M., Zhang, X.H., Chen, J.W., Luo, W., Chen, B.X., Chen, J.H., Liu, H., Li, J., Lai, R.F., Bai, M.Z., Wei, J., Yi, S.K., Wang, H.L., Cao, X.J., Zhou, X.Y., Zhao, Y.H., Wei, K.J., Yang, R.B., Liu, B.N., Zhao, S.C., Fang, X.D., Schartl, M., Qian, X.Q. and Wang, W.M., 2017. The draft genome of blunt snout bream (Megalobrama amblycephala) reveals the development of intermuscular bone and adaptation to herbivorous diet. Giga Sci., 6: 1-13. https://doi.org/10.1093/gigascience/gix039
Li, S.Z. and Wang, H.M., 1987. Osteological studies of some Chinese flatfishes II. The vertebrae, ribs,epipleurals and myoseptal bones. Curr. Zool., 33: 267-276.
Li, Z., Zhou, L., Wang, Z.W., Li, X.Y., Zhang, X.J., Wang, Y. and Gui, J.F., 2017. Comparative analysis of intermuscular bones between clone A+ and clone F strains of allogynogenetic gibel carp. Acta Hydrol. Sin., 4: 860-869.
Lv, Y.P., Bao, B.L., Jiang, Y., Yang, L.L. and Li, J.L., 2007. Comparative analysis of intermuscular bones in lower teleosts. J. Fish. China, 31: 661-668.
Lv, Y.P., Chen, J., Bao, B.L. and Huang, P.P., 2012. The ossificational process of the intermuscular bones in Hemibarbus labeo. J. Shanghai Ocean Univ., 4: 549-553.
Ma, L.X., Dong, Z.J., Su, S.Y., Zhang, J.Q., Liu, W., Li, L.L. and Yuan, X.H., 2012. Research progress of fishes intermuscular bones. J. Jiangsu Agric. Sci., 40: 234-235.
Meng, Q.W., Su, J.X. and Li, W.D., 1987. Comparative anatomy of fish. Science Press, Beijing, China. pp. 102-103.
Nie, C.H., Wan, S.M., Tea, T., Tomislav, T., Hsiao, C.D., Wang, W.M. and Gao, Z.X., 2017. Comparative proteomics analysis of teleost intermuscular bones and ribs provides insight into their development. BMC Genom., 147: 1-14. https://doi.org/10.1186/s12864-017-3530-z
Olson, E.N., 1992. Interplay between proliferation and differentiation within the myogenic lineage. Devel. Biol., 154: 261-272. https://doi.org/10.1016/0012-1606(92)90066-P
Patterson, C. and Johnson, G.D., 1995. The intermuscular bones and ligaments of teleostean fishes. Smiths. Contrib. Zool., 559: 1-85. https://doi.org/10.5479/si.00810282.559
Pownall, M.E., Gustafsson, M.K. and Emerson, C.P. Jr., 2002. Myogenic regulatory factors and the specification of muscle progenitors in vertebrate embryos. Annu. Rev. Cell Devel. Biol., 18: 747-783. https://doi.org/10.1146/annurev.cellbio.18.012502.105758
Quiat, D., Voelker, K.A., Pei, J., Grishin, N.V., Grange, R.W., Bassel-Duby, R. and Olson, E.N., 2011. Concerted regulation of myofiber-specific gene expression and muscle performance by the transcriptional repressor sox6. Proc. natl. Acad. Sci. USA, 108: 10196-10201. https://doi.org/10.1073/pnas.1107413108
Rom, M. and Finkel, A., 1975. Variability of intermuscular bones, vertebrae, ribs, dorsal fin rays and skeletal disorders in the common carp. Theoret. appl. Genet., 46: 33-43. https://doi.org/10.1007/BF00264753
Stickney, H.L., Stickney, Michael, J.F., Barresi, and Stephen, H.D., 2000. Somite development in zebrafish. Devel. Dynam., 219: 287-303. https://doi.org/10.1002/1097-0177(2000)9999:9999<::AID-DVDY1065>3.0.CO;2-A
Sun, W., 2008. Cloning, expression and functional analysis of genes regulating muscle development from zebrafish. PhD thesis, Chinese Academy of Science, Qingdao, China. pp. 10-11.
Watabe, S., 1999. Myogenic regulatory factors and muscle differentiation during ontogeny in fish. J. Fish. Biol., 55: 1-18. https://doi.org/10.1111/j.1095-8649.1999.tb01042.x
Tan X. G., Du S. J., 2002. Differential expression of two myod genes in fast and slow muscles of gilthead seabream (Sparus aurata). Devel. Genes. Evolut., 212: 207-217. https://doi.org/10.1007/s00427-002-0224-5
Ticho, B.S., Stainier, D.Y.R., Fishman, M.C. and Breitbart, R.E., 1996. Three zebrafish mef2 genes delineate somitic and cardiac muscle development in wild-type and mutant embryos. Mech. Devel., 59: 205-218. https://doi.org/10.1016/0925-4773(96)00601-6
Von, H.J., Elworthy, S., Gilchrist, M.J., James, C.S., Fiona, C.W. and Philip, W.I., 2008. Concerted regulation of myofiber-specific gene expression and muscle performance by the transcriptional repressor sox6. EMBO Rep., 9: 683-689. https://doi.org/10.1038/embor.2008.73
Wan, S.M., Yi, S.K., Zhong, J., Nie, C.H., Guan, N.N., Zhang, W.Z. and Gao, Z.X., 2016. Dynamic mRNA and miRNA expression analysis in response to intermuscular bone development of blunt snout bream (Megalobrama amblycephala). Scient. Rep., 6: 1-13. https://doi.org/10.1038/srep31050
Weinberg, E.S., Allende, M.L., Kelly, C.S., Abdelhamid, A., Murakami, T., Andermann, P., Doerre, O.G., Grunwald, D.J. and Riggleman, B., 1996. Developmental regulation of zebrafish myod in wild-type,no tail and spadetail embryos. Development, 122: 271-280.
Yao, W.J., Lv, Y.P., Gong, X.L., Wu, J.M. and Bao, B.L., 2015. Different ossification patterns of intermuscular bones in fish with different swimming modes. Biol. Open, 12: 1727-1732. https://doi.org/10.1242/bio.012856
Yun, K. and Wold, B., 1996. Skeletal muscle determination and differentiation: story of a core regulatory network and its context. Curr. Opin. Cell Biol., 8: 877-889. https://doi.org/10.1016/S0955-0674(96)80091-3
To share on other social networks, click on any share button. What are these?









