Advances in Animal and Veterinary Sciences
Research Article
Prevalence and Virulence Genes of Vibrio and Aeromonas Species Isolated from Nile Tilapia and Mugil Fish Farms in Egypt
Ashraf A. Abd El-Tawab1, Fatma I. El-Hofy1, Gamal R. Hasb-Elnaby2, Manar E. El-Khayat1, Mennat Allah Refaey2*
1Department of Bacteriology, Immunology, and Mycology, Faculty of Veterinary Medicine, Benha University, Egypt; 2Agriculture Research Center, Animal Health Research Institute, Tanta Branch, Egypt.
Abstract | The present study was designed to determine the prevalence of Vibrio and Aeromonas in Nile tilapia and Mugil fish farms in Egypt and monitoring some virulence genes associated with V. parahaemolyticus and A. hydrophila isolates. One hundred diseased fishes (50 for each Nile tilapia and Mullet fish) were collected over 10 months from January to October 2019 and submitted to bacteriological and biochemical examination. Randomly selected 5 isolates of Vibrio species were submitted for molecular identification using species-specific PCR (toxR gene for V. parahaemolyticus and collagenase for V. alginolyticus) and the molecular detection of virulence genes including recA and trh in V. parahaemolyticus isolates. Other 5 A. hydrophila isolates were examined for the molecular detection of virulence genes including fla, aerA, hlyA, and ahcytoen genes. Vibrio species isolated with a prevalence of 65.0% for 100 examined fishes. Vibrio parahaemolyticus, V. alginolyticus, and V. cholerae were isolated with a prevalence of 55.4, 33.8, and 10.8%, respectively. Aeromonas species were isolated with a prevalence of 72.0% for 100 examined fishes where A. hydrophila and A. caviae were isolated with a prevalence of 99.3 and 9.7%, respectively. PCR results showed toxR gene was detected in all five isolated Vibrio species, recA virulence gene was detected in three out of the five tested V. parahaemolyticus isolates. Meanwhile, the trh gene was not detected. On the other hand, aerA and hlyA virulence genes were detected in all five isolated A. hydrophila. Ahcytoen was detected in 4 out of 5. Meanwhile, the fla gene was detected in 1 out of the tested 5 A. hydrophila isolates. The high prevalence of Vibrio and Aeromonas species in this study with an elevated coexistence of their virulence genes threatens the aquaculture industry in Egypt and poses a public health concern.
Keywords | Aeromonas, Vibrio, Nile tilapia, Mugil cephalus, Virulence genes
Received | February 17, 2021; Accepted | June 12, 2021; Published | August 25, 2021
*Correspondence | Mennat Allah Refaey, Agriculture Research Center, Animal Health Research Institute, Tanta Branch, Egypt; Email: [email protected]
Citation | El-Tawab AAA, El-Hofy FI, Hasb-Elnaby GR, El-Khayat ME, Refaey MA (2021). Prevalence and virulence genes of vibrio and Aeromonas species isolated from Nile tilapia and mugil fish farms in Egypt. Adv. Anim. Vet. Sci. 9(10): 1625-1631.
DOI | http://dx.doi.org/10.17582/journal.aavs/2021/9.10.1625.1631
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2021 El-Tawab et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
Aquaculture industry has expanded rapidly worldwide, which offers better economic income, job opportunities, and high-quality food products (Manage, 2018). Egypt is producing about 73.8% of total cultured fish in Africa and occupies the eighth level all over the world as it produces about 919585 tons of cultured fish that represents 1.54% of total cultured fish all over the world (FAO, 2012). Bacterial diseases seriously affect the fish industry, which may be attributed to the high death rate not only of farmed fish but also wild fish (Khalil and Abd El-Latif, 2013). Several Vibrio species are well recognized for their severity to cause fish disease, besides, causing mortality in reared fish is very common during early larval stages and can occur suddenly, leading sometimes to the death of the population (Thompson et al., 2004). The most studied Vibrio spp. known to be pathogenic to humans are Vibrio parahaemolyticus, Vibrio alginolyticus, and Vibrio vulnificus (You et al., 2016). Commonly, not all strains of V. parahaemolyticus are considered pathogenic (Dileep et al., 2003), the pathogenic ones that responsible for the onset of disease symptoms and outbreaks are characterized by the production of thermostable toxins and/or TDH-related hemolysin encoded by tdh and trh genes; respectively (World Health Organization and Food Agriculture Organization, 2011).
Aeromonas is the most prominent pathogen that can infect fish; many Aeromonas species are responsible for several fish diseases including A. hydrophila, A. caviae, and A. veronii biotype sobria. Aeromonas hydrophila infections are characterized by septicemia, ulcerative, hemorrhagic diseases, and significant mortalities in both wild and farmed freshwater and marine fish species. They don’t only cause mortality and cytotoxicity but also create uselessness of live fish as they are responsible for spots, lesions, and scale loss on the infected ones (Saikot et al., 2013). The pathogenicity of aeromonads has been linked to exotoxins that were grouped as aerolysin-hemolysins, cytolytic enterotoxins, or cytotonic enterotoxins (Kingombe et al., 1999). Vibrios and Aeromonas cause a great economic problem for aquaculture and human consumers in Egypt. Therefore, our study aimed for studying the prevalence of these bacteria in fresh Nile tilapia and Mugil cephalus isolated from the different fish farms at Kafr Elsheikh governorate and the molecular detection of some virulence genes.
MATERIALS AND METHODS
Ethical approval
All samples were taken according to the standard sample collection procedure without putting any stress on the fish sample. The current study was approved by the Ethical Committee for Medical Research at the faculty of veterinary sciences, Benha University and Animal Care Guidelines of the General Organization for Veterinary Services, Egypt.
Collection of samples
One hundred (n=100) clinically diseased fish samples,50 Nile tilapia (Oreochromis niloticus) and 50 mullet fish (M. cephalus), complaining of high mortality rate with signs of septicemia (unilateral or bilateral exophthalmia, skin ulcers, and hemorrhages) were collected from different fish farms at Kafr Elsheikh Governorate, Egypt at the period between January (2019) and October (2019). Each examined sample was taken alone in a strong sterile plastic bag with ½ of its volume water pumped with pressured oxygen, labeled and transferred alive with a minimum delay to the laboratory (Microbiology unit of Animal Health Research, Tanta branch, Egypt) for clinical and bacteriological examination. Three hundred and five samples were collected under strict aseptic conditions from apparently pathognomonic lesions in liver, kidney, spleen, heart, intestine, and gills from the 100 diseased fishes; 157 lesion samples from 50 Nile tilapia (O. niloticus) and 148 samples from 50 mullet fish (M. cephalus) and submitted to bacteriological and biochemical examination for isolation of Vibrio and Aeromonas species.
Bacteriological examination
Isolation and identification of Vibrio and Aeromonas species using the conventional cultural method
According to (Quinn et al., 2002) and (Markey et al., 2013) sterilized loops were introduced through seared lesions using a hot spatula. Then taken from lesions and inoculated into 1% peptone broth (Oxoid, UK) + 3%NaCl for Vibrio isolates and 1% peptone broth for Aeromonas isolates then incubated aerobically at 37°C for 18-24 hours. Loops from incubated cultured broth were streaked onto selective diagnostic agar media: cholera medium (TCBS, Oxoid) for Vibrio spp. and Aeromonas selective agar (BSIBG agar, HIMEDIA, M1890-55 G) for Aeromonas spp. and incubated for 24 hours at 37ºC. After that one separated typical colony from agar medium was picked up and transferred into the nutrient broth (Oxoid, UK) and was aerobically incubated at 37 ºC for 18-24 hrs, then 15% glycerol was added, gently mixed, and immediately preserved in a refrigerator at -85 ºC. The smears from the suspected pure colonies were stained with Gram’s stain and microscopically examined under the oil immersion lens. Vibrio spp. (Gram-negative, curved bacilli, comma shape) and Aeromonas spp. (Gram-negative, straight rods with a round end, non-capsulated and non-sporulated) were selected for further biochemical identification steps later. The biochemical identification of Vibrio and Aeromonas isolates was performed according to (Quinn et al., 2002; Nicky, 2004; Markey et al., 2013) by using catalase, oxidase, indole production, citrate utilization, urease test, triple sugar iron, and methyl red tests. Vibrio species were positive for oxidase; catalase, indole production, methyl red, citrate utilization except V. alginolyticus were citrate utilization negative, and ferment the glucose without gas, negative results for urease test and H2S production except V. alginolyticus produced H2S. Aeromonas hydrophila isolates were positive for oxidase, catalase, indole, H2S production, and ferment glucose with gas production. While negative for citrate utilization and methyl red tests. Aeromonas caviae are similar to A. hydrophila except for methyl red +ve, H2S –ve, and not ferment glucose.
Molecular identification of virulence genes in Vibrio and Aeromonas isolates
PCR was used for the confirmation of Vibrio species (5 random isolates) using species-specific genes (toxR gene for V. parahaemolyticus and collagenase gene for V. alginolyticus), as well as the detection of virulence genes of V. parahaemolyticus and A. hydrophila species by primers targeting different virulence genes including recA and trh genes for V. parahaemolyticus as well as aerA, hlyA, ahcytoen and fla genes for A. hydrophila (Table 1).
Extraction of DNA from Vibrio and Aeromonas isolates
It was performed by QIAamp® DNA Mini Kit (Catalogue no. 51304) according to the manufacturer’s instructions.
Amplification and cycling protocol of PCR for Vibrio and Aeromonas isolates
The cycling condition for each gene was prepared according to Emerald Amp GT PCR master mix (Takara, Cat PR310A) kit (Table 1). The amplification was performed on Eppendorf MasterCycler® (Eppendorf AG, Hamburg, Germany) in a total reaction volume of 25 µl containing 12.5 µl EmeraldAmp GT PCR Master Mix, 1 µl of each forward and reverse primers, 4.5 µl molecular biology grade water, and 6 µl test DNA.
Detection of PCR products
The PCR amplicons were analyzed by electrophoresis using a 1.5 % agarose gel in TBE buffer (45 mM Tris-borate, 1 mM EDTA, pH = 8.3). A 100 bp plus DNA Ladder (Qiagen, Germany, GmbH) was used to determine the fragment sizes.
RESULTS AND DISCUSSION
The prevalence of Vibrio and Aeromonas species in diseased fishes
Out of 100 diseased fish samples (50 from each Nile Tilapia and Mullet fish), 65 isolates of Vibrio species were isolated and biochemically identified. A total of 36 V. parahaemolyticus strains (55.4%) were isolated and identified, 20 (30.76%) from O. niloticus, and 16 (24.6%) from M. cephalus. Meanwhile, 22 V. alginolyticus strains (33.8%) were isolated, 13 (20.0%) from O. niloticus, and 9 (13.8%) from M. cephalus fish. Besides 7 V. cholera strains (10.8%) were isolated, 5 (7.7%) from O. niloticus, and 2 (3.1%) from M. cephalus (Table 2).
Table 1: Oligo-nucleotide primers and cycling conditions of the primers used in this study.
| Bacterial spp. | Target genes |
Oligonucleotide sequence (5′ → 3′) |
Product size (base pairs) |
Primary Denaturation |
Amplification (35 cycles) |
Final extension | References | |||
| Secondary denaturation | Annealing | Extension | ||||||||
|
Vibrio alginolyticus |
collagenase | F | CGAGTACAGTCACTTGAAAGCC | 737 |
94˚C 5 min. |
94˚C 1 min. |
50˚C 1 min. |
72˚C 1 min. |
72˚C 10 min. |
|
| R | CACAACAGAACTCGCGTTACC | |||||||||
| Vibrio parahaemolyticus |
ToxR |
F | GTCTTCTGACGCAATCGTTG | 368 |
96˚C 5 min. |
94˚C 1 min. |
63˚C 1.5min. |
72˚C 1.5min. |
72˚C 10 min. |
|
| R | ATACGAGTGGTTGCTGTCATG | |||||||||
|
recA |
F | TGARAARCARTTYGGTAAAGG | 793 |
95˚C 5 min. |
95˚C 35 sec. |
55˚C 1 min. |
72˚C 1 min. |
72˚C 7min. |
||
| R | TCRCCNTTRTAGCTRTACC | |||||||||
| trh | F | ACCTTTTCCTTCTCCWGGKTCSG | 484 |
94˚C 5 min. |
95˚C 1 min. |
62˚C 1 min. |
72˚C 1 min. |
72˚C 2 min. |
||
| R | RCCGCTCTCATATGCYTCGACAKT | |||||||||
| Aeromonas hydrophila | fla | F | TCCAACCGTYTGACCTC | 608 |
94˚C 5 min |
94˚C 30 sec. |
55˚C 40 sec. |
72˚C 45 sec. |
72˚C 10 min. |
|
| R | GMYTGGTTGCGRATGGT | |||||||||
|
aerA |
F | CACAGCCAATATGTCGGTGAAG | 326 |
94˚C 3 min. |
94˚C 30 sec. |
52˚C 30 sec. |
72˚C 30 sec. |
72˚C 10 min. |
||
| R | GTCACCTTCTCGCTCAGGC | |||||||||
|
hlyA |
F | GGCCGGTGGCCCGAAGATACGGG | 592 |
95˚C 5 min. |
95˚C 2 min. |
55˚C 1 min. |
72˚C 1 min. |
72˚C 7min. |
||
| R | GGCGGCGCCGGACGAGACGGGG | |||||||||
| Ahcytoen | F | GAGAAGGTGACCACCAAGAACAA | 232 |
94˚C 5 min. |
94˚C 30 sec. |
56˚C 30 sec. |
72˚C 1 min. |
72˚C 10 min. |
||
| R | AACTGACATCGGCCTTGAACTC | |||||||||
Table 2: Prevalence of Vibrio species isolated from examined fishes.
| Type and number of fish samples | No. of examined fish samples | No. of examined lesion samples |
Positive samples for Vibrio species |
Total | ||||||
| V. parahaemolyticus | V. alginolyticus | V. cholerae | ||||||||
| No. | %* | No. | %* | No. | %* | No. | %* | |||
|
Nile tilapia (O. niloticus) |
50 | 157 | 20 | 30.7 | 13 | 20.0 | 5 | 7.7 | 38 | 58.5 |
|
Mullet (M. cephalus) |
50 | 148 | 16 | 24.6 | 9 | 13.8 | 2 | 3.1 | 27 | 41.5 |
| Total | 100 | 305 | 36 | 55.4 | 22 | 33.8 | 7 | 10.8 | 65 | 100 |
*Percentage in relation to the total number of Vibrio species isolated (65).
Table 3: Prevalence of Aeromonas species isolated from examined fishes.
| Type and number of fish samples | No. of the examined fish samples | No. of examined lesion samples |
Positive samples for Aeromonas species |
|||||
| A. hydrophila | A. caviae | Total | ||||||
| No. | %* | No. | %* | No. | %* | |||
|
Nile tilapia (O. niloticus) |
50 | 157 | 26 | 36.1 | 1 | 1.4 | 27 | 37.5 |
|
Mullet (M. cephalus) |
50 | 148 | 39 | 54.2 | 6 | 8.3 | 45 | 62.5 |
| Total | 100 | 305 | 65 | 90.3 | 7 | 9.7 | 72 | 100.0 |
*Percentage in relation to the total number of Aeromonas species isolated (72).
Table 4: Types and prevalence of virulence genes in V. parahaemolyticus and A. hydrophila strains.
|
V. parahaemolyticus |
Aeromonas hydrophila |
||||||||||
|
recA |
trh | fla |
aerA |
hlyA |
ahcytoen | ||||||
| No. | % | No. | % | No. | % | No. | % | No. | % | No. | % |
| 3 | 60.0 | 0 | 0.0 | 1 | 20.0 | 5 | 100.0 | 5 | 100.0 | 4 | 80.0 |
A total of 72 Aeromonas species were isolated and identified. Sixty-five A. hydrophila strains (90.3 %) were isolated, 26 (36.1%) from O. niloticus and 39(54.2%) from M. cephalus. Meanwhile, 7 A. caviae strains (9.7 %) were isolated, 1 (1.4%) from O. niloticus and 6 (8.3%) from M. cephalus (Table 3).
Molecular identification of Vibrio species using PCR
As a result of the molecular screening of 5 Vibrio species isolates using species-specific PCR (for V. parahaemolyticus (toxR gene) which generated at 368 bp and for V. alginolyticus (collagenase gene) which generated at 737bp), all five isolates were identified as V. parahaemolyticus. On the other hand, no V. alginolyticus isolates were identified (Figure 1).
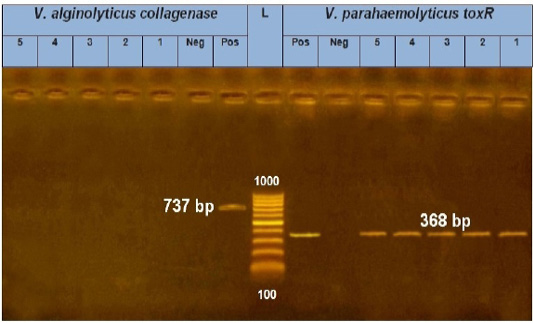
Figure 1: PCR amplification of the toxR gene (V. parahaemolyticus) and collagenase gene (V. alginolyticus) on agarose gel 1.5%. Lane L: 100-1000 bp. DNA Ladder. Neg.: Negative control (Enterobacteriaceae strain). Pos.: Positive control (local strain obtained from Central Lab. for quality control of poultry production, El-Giza, Egypt (CLQP)) (at 368 bp for toxR gene and 737bp for collagenase gene). Lanes 1-5: V. parahaemolyticus (toxR gene) positive and Lanes 1-5: V. alginolyticus (collagenase gene) negative.
Prevalence of some virulence genes among some isolated V. parahaemolyticus and A. hydrophila using PCR
PCR results showed the recA virulence gene was detected in three out of five random isolated V. parahaemolyticus. Meanwhile, the trh virulence gene was not detected in any of the tested 5 V. parahaemolyticus isolates (Figure 2). Also, five randomly selected A. hydrophila isolates were submitted for the screening of virulence genes by PCR. Results revealed that aerA and hlyA virulence genes were detected in all five random isolated A. hydrophila (Figure 3), Meanwhile, Ahcytoen was detected in 4 out of 5 A .hydrophila studied strains and fla virulence gene was detected in 1 out of 5 A. hydrophila isolates (Figure 4) (Table 4).
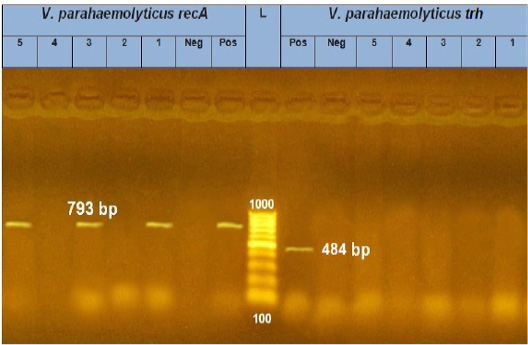
Figure 2: PCR amplification of recA and trh genes of V. parahaemolyticus on agarose gel 1.5%. Lane L: 100-1000 bp. DNA Ladder, Neg.: Negative control (Enterobacteriaceae strain), Pos.: Positive control (local strain obtained from CLQP) (at 793 bp for recA gene and 449 bp for trh gene), Lanes 1, 3 and 5: V. parahaemolyticus (recA gene) positive, Lane 2 and 4: V. parahaemolyticus (recA gene) negative, and Lanes 1-5: V. parahaemolyticus (trh gene) negative
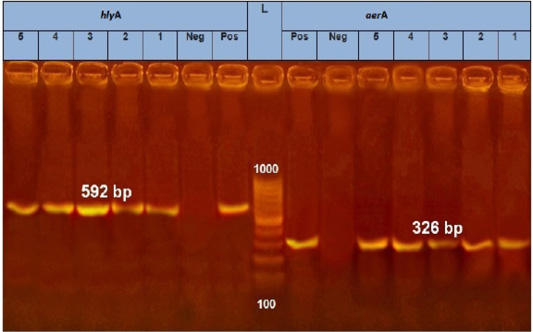
Figure 3: PCR amplification of Aerolysin (aerA) gene and hemolysin (hlyA) on agarose gel 1.5%. Lane L: 100-1000 bp. DNA Ladder, Neg.: Negative control (Enterobacteriaceae strain), Pos.: Positive control (local strain obtained from CLQP) (at 326 bp for aerA gene and 592 for hlyA gene), Lanes 1-5: A. hydrophila (aerA gene) positive and Lanes 1-5: A. hydrophila (hlyA gene) positive.
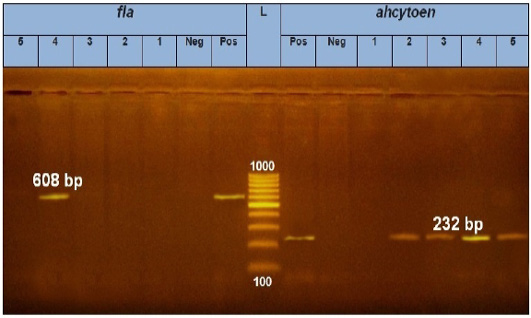
Figure 4: PCR amplification of fla and ahcytoen genes on agarose gel 1 %. Lane L: 100-1000 bp. DNA Ladder. Neg.: Negative control (Enterobacteriaceae strain), Pos.: Positive control (local strain obtained from CLQP) (at 608bp for fla gene and 232bp for ahcytoen gene, Lane 4: A. hydrophila (fla gene) (Positive), Lanes 1, 2, 3and 5: A .hydrophila (fla gene) (Negative), Lanes 2, 3, 4 and5: A. hydrophila (ahcytoen gene) (Positive) and Lane 1: A. hydrophila (ahcytoen gene) (Negative).
Discussion
The bacterial infection is one of the main obstacles to aquaculture, which leads to wide economic losses (Gophen, 2017). This study was planned for the determination of the prevalence of Vibrio and Aeromonas infection in Nile tilapia and Mullet fish farms and the molecular detection of some virulence genes. The obtained results of naturally infected fishes with Vibrio species indicating disease problem and the clinical signs varied from the dark coloration of skin with detached scales, and hemorrhage at the base of the fins with some erosion which support the findings of El-Bouhy et al. (2016), and Al-Taee et al. (2017) in Mugil and Tilapia. Vibrio parahaemolyticus was the most highly detected with a prevalence of 55.4 % and detected in Nile tilapia higher than mullet fish (Table 2) which may be due to the flesh texture of tilapia fish or their living habits. These results were in agreement with that obtained by El-Hady et al. (2015). The reverse result obtained by Abdelaziz et al. (2017) where V. alginolyticus recorded the highest prevalence of infection followed by V. parahaemolyticus is probably due to differences in fish species (Solea senegalensis and Mugil capito). Lower prevalence of V. parahaemolyticus was recorded by Abdelaziz et al. (2017); Ahmed et al. (2018), and Deng et al. (2020) which may be due to differences in the geographical origin or season. Five random Vibrio species isolates were further genetically verified by PCR for the detection of V. parahaemolyticus toxR gene. Kim et al. (1999) clarified that the V. parahaemolyticus toxR sequence is perfectly conserved among V. parahaemolyticus strains and always be an appropriate diagnostic test. In our study, all five isolates were identified as positive for the toxR gene as V. parahaemolyticus (Figure 1). These results agreed with that recorded by Al- Othrubi et al. (2014) and Ashrafudoulla et al. (2019) where the toxR gene was detected in all V. parahaemolyticus isolates. PCR results showed that the trh virulence gene was not detected in any of the tested 5 V. parahaemolyticus isolates indicating that the majority of isolates were found to be non-pathogenic to humans due to the lack of the pathogenic trh gene (Figure 2). These results agreed with that recorded by Mohamad et al. (2019) and Tan et al. (2020). Meanwhile, the recA virulence gene was detected in three out of five tested V. parahaemolyticus isolates (Figure 2). Theethakaew et al. (2013) identified frequent interspecies horizontal gene transfer and intra genetic recombination at the recA locus, which plays a great role in the apparent evolutionary classification of V. parahaemolyticus (Han et al., 2014). Aeromonas species cause severe diseases among fish and humans, as Motile Aeromonas Septicemia (MAS) in fish which is caused by A. hydrophila leading to high mortalities and high economic losses (Shayo et al., 2012). The results of clinical and postmortem examinations of studied fish were similar to those reported by Sayed (2017) and Algammal et al. (2020). The results of bacteriological examination (Table 3) revealed that A. hydrophila was isolated in mullet fish higher than Nile tilapia. These results agreed with these of Abd El Tawab et al. (2017), and Enany et al. (2019). Lower results were recorded by Gufe et al. (2019), and Algammal et al. (2020) which was probably due to differences in the geographical distribution. The aerA gene was amplified in all five random isolated A. hydrophila studied strains (Figure 3). These results were similar to Algammal et al. (2020), and Sonkol et al. (2020) who reported that the aerolysin (aerA) gene was detected in 100% of the isolates. Meanwhile, they have disagreed with that recorded by Nagar et al. (2011) and Ibrahim (2015) who failed to detect aerA virulent gene in these strains. The hlyA gene was amplified in all five random isolated A. hydrophila studied strains (Figure 3). Similar results were decided by Ruhil-Hayati et al. (2015) and Abd El Tawab et al. (2017). Lower results were reported by Sonkol et al. (2020). The Ahcytoen gene was amplified in 4 out of 5 A. hydrophila studied strains (Figure 4). Similar results were recorded by Abd El Tawab et al. (2017). The fla gene was amplified in 1 out of 5 A. hydrophila studied strains (Figure 4). Similar results were reported by Blaszk (2013) and Aravena et al. (2014). While higher results were reported by Nawaz et al. (2010) and Dahanayake et al. (2020).
CONCLUSIONS AND RECOMMENDATIONS
This study revealed that Vibrio and Aeromonas species contribute to the occurrence of severe economic losses in the aquaculture industry in Egypt that requires a constant detection of these isolates to reduce the spread of infection. The high frequency of virulence genes in the isolates obtained in this work revealed the important role of these virulence genes in the emergence of the clinical signs.
ACKNOWLEDGEMENTS
The authors appreciate the members of the Animal Health Research Institute, Tanta Branch, Egypt for their encouragement during the working and examination of the samples. We thank all staff members of the Department of Bacteriology, Immunology and Mycology, Faculty of Veterinary Medicine, Benha University.
Author’s Contribution
All authors contributed equally.
Conflict of interest
The authors have declared no conflict of interest.
REFERENCES






