Advances in Animal and Veterinary Sciences
Research Article
Molecular Identification and Phylogenetic Analysis of Theileria equi and Babesia caballi Infections in Equids from Erbil Province, North of Iraq
Khalid Jabar Aziz1*, Lokman Taib Omer AL-Barwary2, Zeravan Abdulrazaq Mohammed3, Ibrahim Abdulqader Naqid4
1Department of Animal Resources, College of Agricultural Engineering Sciences, Salahuddin University, Erbil; 2Department of Microbiology and Pathology, College of Veterinary Medicine, University of Duhok; 3 Duhok Research Center, College of Veterinary Medicine, University of Duhok; 4Department of Biomedical Science, College of Medicine, University of Zakho
Abstract | This study performed molecular detection and analysis of the heterogeneity of the 18S rRNA gene isolates obtained from equids such as horse, mule, donkey and pony from Erbil province, Kurdistan region of Iraq. Multiplex polymerase chain reaction (PCR) indicated that 76/136 (55.88%) of equine were infected with piroplasms, with Theileria equi (P=41.91%; CI=3.76-14.77%) more prevalent than Babesia caballi (P=8.82%; CI=1.00%), while mixed infection was less (P=5.15%; CI=0.21-1.46%) with a significant difference (P<0.001). There was a significant association between the prevalence of T. equi and recreation (p<0.03) or racing (p<0.02), and but neither the type of equids nor the gender and age groups was significantly associated with prevalence. The obtained sequences were utilized for characterizing the genotypes and phylogeny of both the protozoa. BLAST analysis indicated 98-100% similarity to species isolated in Turkey, Malaysia, Egypt, Sudan, Jordan, Iran, Brazil and South Africa. Four 18S rRNA genotype clades were observed for T. equi (A, B, C and D) and two for B. caballi (A and B). Genetic variation found among the equids in Erbil province is probably due to introduction of equines from other countries without quarantine measures. This study indicates that infection with T. equi is more prevalent than that of B. caballi in the studied area.
Keywords | Multiplex PCR, Phylogenetic analysis, Equine piroplasmosis, Erbil
Received | September 26, 2019; Accepted | November 10, 2019; Published | November 20, 2019
*Correspondence | Khalid Jabar Aziz, Department of Animal Resources, College of Agricultural Engineering Sciences, Salahuddin University, Erbil; Email: [email protected]
Citation | Aziz KJ, AL-Barwary, LTO, Mohammed ZA, Naqid IA (2019). Molecular identification and phylogenetic analysis of theileria equi and babesia caballi infections in equids from Erbil Province, North of Iraq. Adv. Anim. Vet. Sci. 7(12): 1060-1066.
DOI | http://dx.doi.org/10.17582/journal.aavs/2019/7.12.1060.1066
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2019 Aziz et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
Equine piroplasmosis (EP) is one of the most common and vital tick-borne diseases and is caused by two hemoprotozoa species, specifically T. equi and B. caballi (Wise et al., 2014). It affects equids including horses, mules, donkeys, pony and zebras, in which it causes substantial direct and indirect losses (Al-Obaidi et al., 2014; Scoles and Ueti, 2015; Del Pino et al., 2016; Aziz and Al-Barwary, 2019). T. equi and B. caballi are transmitted by ixodid ticks of the genera Hyalomma, Rhipicephalus and Dermacentor (de Waal and Heerden, 1994). Their infections which are indicated by an acute, sub-acute or chronic clinical sign, with mortality rate of up to 50% (de Waal, 1992).
The distribution of EP is worldwide. It is particularly prevalent in tropical and subtropical areas. It is endemic in many parts of Asia, Africa, Europe, Arabia and America (Leblond et al., 2005). The clinical symptoms of EP are often varied and non-specific. Therefore, distinction between both type of infection based on clinical signs alone is impossible and mixed infections also occur (Potgieter et al., 1992; Rothschild and Knowles, 2007).
Definitive diagnosis of equine piroplasmosis depends on the demonstration of B. caballi and T. equi in blood smears stained with Giemsa stain (Ali et al., 1996). However, the percentages of parasitemia in carrier state of less than 0.1% infected erythrocytes are not detected by Giemsa stain blood smears (Seifi and Sardari, 2000; Krause, 2003). The serological assessments also do not distinguish between existing and previous infection (Holman et al., 1997).
In order to come up with better diagnosis of many species of Babesia and Theileria and with the aim of obtaining higher sensitivity and specificity compared to other serological diagnostic methods, PCR has been applied (Buling et al., 2007; Jefferies et al., 2007; Sibeko et al., 2008).
PCR assay has also been recommended for the detection of latent infection of both infections and can also be used to identify the genetic diversity and phylogenetic relationships of rising Theileria and Babesia species (Schnittger et al., 2004; Moretti et al., 2010). Besides, use of molecular tools for identification and detection of a number of strains and species would permit higher insight into the genetic diversity of equine piroplasms present in our region. Therefore, the present study aimed to evaluate the incidence of EP through the use of molecular tools and document the phylogenic diversity of T. equi and B. caballi among different species of equids based on 18S rRNA sequences for the first time in Kurdistan region, Iraq.
MATERIALS AND METHODS
Study area and sampling
This study was carried out under the supervision and regulations of Ethic Committee at College of Veterinary medicine, University of Duhok, Iraq. 136 equids (85 horses, 27 mule, 19 donkey and 5 ponies) were sampled from April 2016 to March 2017 in various geographic areas of Erbil province, Iraq. An 18-sized needle was used to collect blood samples from the jugular vein and were placed into sterile vacutainers® tubes (5 ml each) with anticoagulant ethylenediaminetetraacetic acid (EDTA). The samples were transported under refrigeration to the laboratory and stored at −20ºC for their molecular analysis.
DNA extraction and PCR amplification
Genomic DNA was extracted from 136 equine blood samples using the Prime Prep TM Genomic DNA Isolation Kit (Genet Bio, South Korea) according to the manufacture’s method. Two types of reactions were achieved: firstly, for identification the positive infection with all possible Theileria spp. and Babesia spp. the universal ‘catch-all’ primers (Bec-UF1and Bec-UR) by conventional PCR assay were used. Secondly, for differentiation between T. equi and B. caballi in all positive and negative samples in the first reaction, two reverse primers and a single forward primer (Bec-UF2) were used, one for identification of T. equi and another for B. caballi, which were respectively Equi-R and Cab-R by multiplex PCR assay (Table 1).
In order to detect both protozoa (T. equi and B. caballi) a multiplex PCR was performed according to the method proposed by Alhassan et al. (2005) and Qablan et al. (2013) with some modifications. Briefly, 25 µl of a mixture containing 2 µl of template DNA, 1 µl (10 pmol) of each reverse primer (EquiR) for T. equi and (CabR) for B. caballi 2 µl of a universal forward primer (UF2), 6.5 dH2O and 12.5 µl of 2X PCR master-mix ready to use (GeNet Bio. Laboratory, Korea) with a final concentration of 0.5 mM of each dNTP in 10 mM Tris-HCl, pH 9.0, 4 mM MgCl2, enzyme stabilizer, loading dye and 1 U Taq DNA polymerase. A BioRad thermocycler was used to subdue the mixtures to the following cycling conditions: 96ºC for 10 minute, with 35 cycle of a denaturing step at 96ºC for 1 minute, an annealing step at 54ºC for 1 minute, and an extension step at 72ºC for 1 minute, followed by a final extension at 72ºC for 10 minute. Afterwards the products were stored at 4ºC until removal.
After amplification, gel electrophoresis was run in a 1.5% agarose. 1X TBE buffer was utilized for a 5 µl of the amplificons to be run, immersed in safe dye tank for 45 minute, then visualised by UV illuminator (Proxima 2500 Isogene Life science, Netherland) and the distinct bands of T. equi (392 bp) and B. caballi (540 bp) were detected. DNA positive controls for T. equi and B. caballi were prepared from the blood of infected horses with clinical piroplasmosis which were confirmed by nucleotide sequencing and the sequences submitted to Genebank with accession number MH047214 and MH047215 respectively. Further, DNA extracted from a piroplasm-free horse was utilized as a negative control for amplification of PCR.
DNA sequencing and Phylogenic analysis
In the current study, 22 PCR amplicons (16 for T. equi and 6 for B. caballi) that were positive using multiplex PCR were sent to the commercial company for purification and sequencing (Macrogen Inc. South Korea).
The sequences similarity analyses with previous sequences published in Genbank were performed by using the BLAST programme (http://www.ncbi.nlm.nih.gov/BLAST).
The phylogenic analysis included 13 samples that represent in Erbil province. MEGA7 version (http:// www.megasoftware.net, July 2016) was employed for multiple sequence alignment. The Muscle software was used to align the sequences with 18S rRNA gene of T.equi and B. caballi sequences, derived from GenBank (Edgar, 2004).
Table 1: The oligonucleotide primers used to amplify the hemoparasites 18S rRNA genes of piroplasms. (Alhassan et al., 2005).
| Primers | Sequences 5’-3’ | Target gene | Expected size (bp) |
| Bec-UF1 |
5’-GTTGATCCTGCCAGTAGTCA-3’ |
“Catch-all” (Babesia spp. and Theileria spp.) |
876 Babesia spp |
| Bec-UR |
5’-CGGTATCTGATCGTCTTCGA-3’ |
913 Theileria spp |
|
| Bec-UF2 |
5’-TCGAAGACGATCAGATACCGTCG-3’ |
Specific primers for B. caballi and T. equi |
------- |
| Cab-R |
5’-CTCGTTCATGATTTAGAATTGCT-3’ |
540 | |
| Equi-R |
5’-TGCCTTAAACTTCCTTGCGAT-3’ |
392 |
Bec-UF1 and Bec-UF2: catch-all forward primers; Bec-UR: catch-all reverse primers; Cab-R: B. caballi-specific reverse primer, Equi-R: T. equi-specific reverse primer.
Table 2: Molecular detection rate of Theileria equi and Babesia caballi in Equine by multiplex PCR
| Type of protozoa | No. of equine tested | Multiplex PCR | ||||
| No. positive | Positive % | 95% CI | Odds ration | P value | ||
| B. caballi | 12 | 8.82 | 1.00 | |||
| T. equi | 136 | 57 | 41.91 | 3.76-14.77 | 7.46 | <0.001 |
| Both protozoa | 7 | 5.15 | 0.21-1.46 | 0.56 | 0.24 | |
| Overall | 76 | 55.88 | ||||
Three different algorithms such as maximum likelihood, neighbor-joining and yielded topology-similar results (Tamura-Nei model) were utilized to generate a phylogenetic tree. To estimate the reliability of node 1000 bootstrap replicates were performed.
Statistical analysis
The χ2 and Fisher’s exact test were used to differentiate the prevalence of equine piroplasmosis. Binomial logistic regression in GenStat 12th Edition was used to determine odds ratio and the incidence of T. equi, B. caballi and mixed infection. All candidate variables were kept in the model with significant attributes at P <0.05.
Results
Multivariable analysis for prevalence of EP by Multiplex PCR
Multiplex PCR analysis of 136 DNA samples from the different area of Erbil province showed that 55.88% (76/136) of symptomatic equines were infected with piroplasms (Table 2). The infection rate of T. equi, B. caballi and mixed infection was 41.91%, 8.82% and 5.15% respectively.
Four variable conditions were analysed in this molecular study including the type of equids, gender, age and purpose of keeping (Table 3). The prevalence of T. equi, B. caballi and mixed infection were not significantly different between types of equids, gender and age groups. It also should be noted that recreation (P=0.03) and racing (P=0.02) appeared to be only significant influence for the prevalence of T. equi in this study (Table 3).
PCR amplification analysis
The results of amplified PCR product using universal ‘catch-all’ primers (Bec-UF1and Bec-UR) showed that the DNA band sizes for the first reaction was 876 bp and 913bp, meaning that the samples were positive for Babesia spp. and Theileria spp. respectively (Figure 1). While for the second reaction using single forward primer (Bec-UF2) and two reverse primers, one of them (Cab-R) was specific for B. caballi and another (Equi-R) for T. equi. The DNA band sizes were 392 bp and 540 bp, meaning that the samples were positive for T. equi and B. caballi, respectively (Figure 2).
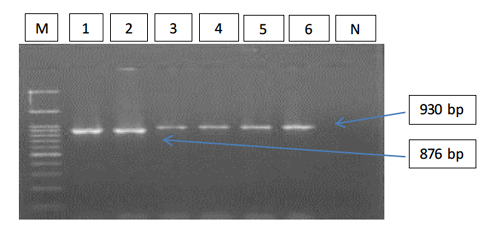
Figure 1: Gel electrophoresis image showing; PCR detection of B. caballi and T. equi with a pair of universal screening primers (Bec-UF1 and Bec-UR): Lanes M) 100 bp ladder DNA marker; Lane 1-2) B. caballi in approximately band size 876bp; lane 3-6) T. equi in approximately band size 930bp; Lane N) negative control.
Phylogenic analysis
T. equi and B. caballi sequences for the hypervariable V4 region of the 18S rRNA gene were obtained from 8 and 5 samples respectively. The sequences were deposited in Genbank with subsequently employed for the phylogenic analysis.
Table 3: Prevalence and relative risk of equids factors associated with multiplex PCR of T. equi, B. caballi and both protozoa.
| Factor | No. of equine tested | T. equi |
|
B. caballi |
|
Both protozoa | ||||||
| N. (%) | OR (95%CI) | p | N. (%) | OR (95%CI) | p | N. (%) | OR (95%CI) | p | ||||
| Type of equine | ||||||||||||
| Donkey | 19 | 9 | 1 | 2 | 1 | 1 | ||||||
| Horse | 85 | 33 | 0.71 (0.26-1.92) | 0.49 | 6 | 0.65 (0.12-3.47) | 0.61 | 3 | 0.65 (0.06-6.06) | 0.72 | ||
| Mule | 27 | 13 | 1.03 (0.32-3.43) | 0.95 | 3 | 1.06 (0.15-7.06) | 0.95 | 2 | 1.44 (0.12-17.04) | 0.77 | ||
| Pony | 5 | 2 | 0.74 (0.10-5.49) | 0.77 | 1 | 2.12 (0.15-29.56) | 0.57 | 1 | 4.50 (0.23-87.65) | 0.32 | ||
| Gender | ||||||||||||
| Female | 58 | 21 | 1 | 3 | 1 | 2 | 1 | |||||
| Male | 51 | 23 | 1.44 (0.67-3.12) | 0.35 | 6 | 2.44 (0.58-10.30) | 0.22 | 3 | 1.75 (0.29-10.67) | 0.55 | ||
| Age group | ||||||||||||
| <5 | 54 | 25 | 1 | 5 | 1 | 3 | 1 | |||||
| 5--10 | 51 | 21 | 0.86 (0.32-2.03) | 0.65 | 4 | 0.83 (0.28-2.45) | 2 | 0.69 (0.21-2.25) | 0.69 | |||
| >10 | 31 | 11 | 0.64 (0.22-1.88) | 0.41 | 3 | 1.05 (0.33-3.43) | 2 | 1.17 (0.35-3.86) | 0.89 | |||
| Purpose of keeping | ||||||||||||
| Breeding | 27 | 5 | 1 | 3 | 1 | 1 | 1 | |||||
| Recreation | 29 | 15 | 4.71 (1.52-19.30) | 0.03 | 1 | 0.29 (0.05-1.71) | 0.17 | 1 | 0.93 (0.16-5.52) | 0.94 | ||
| Racing | 29 | 16 | 5.41 (1.32-22.21) | 0.02 | 4 | 1.28 (0.37-4.45) | 0.69 | 3 | 2.99 (0.68-13.15) | 0.14 | ||
| loading | 51 | 21 | 3.08 (0.84-11.30) | 0.09 | 4 | 0.68 (1.99-2.33) | 0.53 | 2 | 1.06 (0.23-4.99) | 0.94 | ||
These sequences when analysed by BLASTn shared identity ranging from 98-100% with T. equi and B. caballi sequences previously reported. The sequences were clustered in four different groups, A, B, C and D for T. equi while in tow groups A and B for B. caballi.
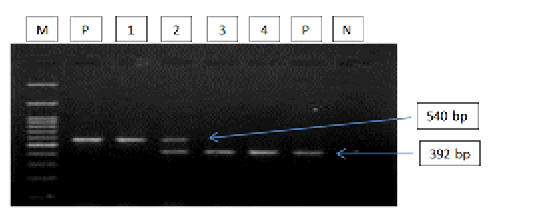
Figure 2: Gel electrophoresis image showing: lane M) 100bp DNA ladder; Lane P) positive control for T. equi and B. caballi; Lane1-4) multiplex PCR detected T. equi and B. caballi in approximately band size 392 and 540 bp respectively; Lane N) negative control.Section of intestine showing sloughing of villli and heterophillic infiltration)
The T. equi tree indicates four major clades representing the genotype (A, B, C and D) (Figure 3). Two sequences (MH368324 and MH368330) detected in horses were clusterd with sequences previously detected from Iran (KJ418423), India (LC042536), South Africa (EU642508) and Brazil (MG052902) belonging to the clade A. Two sequences (MH368315, MH368343) were posisioned near to sequences previously detected from Turkey (KU921663), Sudan (AB515315) and Jordan (KX227637), belonging to the clade B.

Figure 3:Rooted phylogenic tree of Theileria equi as interfered from partial sequences of 18S rRNA gene. Black diamond indicated sequences obtained from the present study; others represent sequences from Genbank.
Two other sequences (MH368338, MH368322) were posisioned near to sequences peviously detected from Jordan (KX227638), Brazil (KJ573373), (MG052917), Iran (KF704750) and Egypt (MF192854) belonging to the clade C.
Finally, two sequences (MH368316 and MH368345) were clustered with Iraq (MH482936), Brazil (KX722525), (MG052915) and Cuba (KY111760), belonging to the clade D.
Tree of B. caballi shows two major clades representing the known genotypes of B. caballi with the exception of genotype C. Four sequences (MH047214, MH368314, MH368304 and MH368302) were placed near the sequences that were previously detected from Jordan (JQ417262), Malaysia (KX349895) and South Africa (EU888904), (Z15104) and (EU642513) belonging to the clade A.
While one sequence (MH368307) was clustered with other from Iran (KM882893), Brazil (KY952238), Central Balkan (KR527221), Turkey (KP792452) and Rumania (KJ908940), belonging to the clade B (Figure 4).

Figure 4:Rooted phylogenic tree of Babesia caballi as interfered from partial sequences of 18S rRNA gene. Black diamond indicated sequences obtained from the present study; others represent sequences from Genbank.
DISCUSSION
Equine piroplasmosis has been previously reported in Iraq using several direct and indirect methods that detected B. caballi and T. equi including Giemsa stained blood smears (Alssad, 2009), cELISA (Alsaad and Al-Obaidi, 2012; Aziz and Al-Barwary, 2019). However these methods are suitable only for the detection in the acute stage of infection, but they are not applicable in identifying pre-symptomatic and carrier animals with low parasetemia (Lobanov et al., 2018). Therefore, this study aimed to investigate the frequency rate of piroplasms infection in different species of equids from Erbil province, North Iraq by molecular technique and to document the phylogenic sequencing of PCR products for the first time. Molecular techniques, as very sensitive and very specific diagnostic tools, have been widely applied to detect and differentiate equine piroplasmosis particularly in carrier animals (Bashiruddin and Rebêlo, 1999; Alhassan et al., 2005).
In this study, molecular examination of equine blood samples obtained from different districts in Erbil province, Iraq, demonstrated that 55.88% (76/136) of symptomatic equines were infected with piroplasms. While the prevalence rate of infection was 41.91% for T. equi and 8.82% for B. caballi. The relatively high prevalence rate of EP observed in the present study could be due to the abundant tick vector population. A previous study conducted in Jordan reported that the prevalence rates of infections were 18.8% for T. equi and 7.3% for B.caballi (Qablan et al., 2013). Another study demonstrated that the infection rates were 4.93% in Turkey; 2.96% and 1.97% for T. equi and B. caballi respectively (Kizilarslan et al., 2015).
Our data show that the infection rate of T. equi in equids by conventional PCR was statistically more significant than that of B. caballi (P<0.05). These findings are in agreement with reports by other authors from horses and donkeys from Iraq and neighbouring countries using different techniques (Alsaad and Al-Obaidi, 2012; Abedi et al., 2014; Kizilarslan et al., 2015). This situation is likely to be caused by active treatment and host immune system leading to efficient elimination of B. caballi, in contrast to T. equi, which has long-life persistence (Bruning, 1996; Laus et al., 2015).
A study of risk factors for EP identified in this study is presented in Table 2. There were no significant differences among the different equids, gender and age groups indicating that EP is widespread in Erbil governorate. T. equi was significantly higher in recreation (p=0.03) and racing (p=0.02) equines. These results are in agreement with Bahrami et al. (2014). This may be due to that physical stressor that may temporarily the immune system and immune-compromised animals have been shown to be more susceptible to infection (Sevinc et al., 2008).
The 18S rRNA gene is broadly employed use for molecular detection and phylogenic analysis of piroplasms in equids (Bhoora et al., 2009; Hall et al., 2013). Here in the molecular detection and phylogenic analysis of T. equi and B. caballi sequences amplified from equids from Erbil province were assessed and the study appears to be the first of its kind from this area.
The current study provides the first information on the genetic diversity of piroplasms in equids population from Iraq based on 18S rRNA gene sequences. Until today, five genotypes of T. equi and three genotypes of B. caballi lineages were established in equine populations from different countries (Bhoora et al., 2010; Salim et al., 2010; Qablan et al., 2013). However by finding a new sequence from various geographical area, further increase in the genetic diversity of T. equi and B. caballi has been predicted (Bhoora et al., 2009; Qablan et al., 2013). In this study, six distinct main clades were identified for T. equi and B. caballi sequences clustering within four and two clades respectively. These findings are in agreement with that reported by Qablan et al. (2013), Kizilarslan et al. (2015) and Ketter-Ratzon et al. (2017). This variation in distribution of piroplasms 18S rRNA gene sequences may be related to the horse and mule business movement from other countries such as Turkey, Iran and Jordan.
The T. equi isolates in our study cluster within genotypes (A, B, C and D), and this may indicate that different genotypes within the same population of equids and tick vectors overlaped, or that a new variant would require further confirmation (Qablan et al., 2013; Salim et al., 2013; Liu et al., 2016).
The B. caballi isolates obtained in the current study clustered within the genotype B and A corresponding to B. caballi that was previously isolated from Jordan (JN596979) (Qablan et al., 2012), Mongolia (JQ288735) (Nakayima, 2015) and South Africa (EU888901) (Bhoora et al., 2009). This variation can be explained based on the mutation in the 18S rRNA gene, which can be benefical to the parasite by the way of revise in the gene expression, which encourage their habituation to a new climate such as a different host (equids and ticks) and environmental changes (Bhoora et al., 2009; Kouam et al., 2010).
In conclusion, T. equi was found to have a high prevalence in equids in Erbil province. This considerable diversity in 18S rRNA found in T. equi may be related to the unregulated equine trade with neihbouring countries. The import of horses into Kurdistan region of Iraq has increased in the last decade. These results support the development of more diagnostic tools for detecting all different genotypes of T. equi and B. caballi in other animals in order to minimise the risk of importing carrier equids.
Further studies on equine piroplasmosis should be more focused on aspects related to the vector (ticks), and this will be an essential component in the study of epidemiology in EP using advanced molecular tools.
ACKNOWLEDGEMENTS
We want to thank Nazhad H. Qader and Yunis A. Ahmad and the farmers for their assistance with the sample collection.
We would also like to thank Miqdad S. Ahmed (Duhok Research Center/Duhok University) and Muhsin J.Abdulwahid (Scientific Research Center/ Salahaddin University) for their help to facilities using laboratory of molecular biology.
Authors contribution
All authors contributed equally.
conflict of interset
The authors declare there is no conflict of interset.
REFERENCES






