Advances in Animal and Veterinary Sciences
Research Article
Quantitative Flow Cytometry Assessment of Feline Circulatory Breast Cancer Stem Cells
Rasha H. Elsabagh 1,2, Haithem A. M. Farghali3*, Ibrahim A. Emam3, Eman Ragab2, Abdelfattah A. Nada4, Salah A. Selim2*
1Animal Health Research Institute (AHRI), Shibin El-Kom Lab, Agriculture Research Centre, 32518 El-Monofiya, Egypt; 2Department of Microbiology, Faculty of Veterinary Medicine, Cairo University, 12211 Giza, Egypt; 3Department of Surgery, Anesthesiology and Radiology, Faculty of Veterinary Medicine, Cairo University,12211 Giza, Egypt; 4Animal Health Research Institute (AHRI), Agriculture Research Centre, 12618 Dokki, Egypt.
Abstract | The upsurge need for a rapid and accurate prognostic system for feline mammary neoplasia is crucial, as almost 90% of mammary tumors in cats tend to metastasize. Here, appears the importance of circulatory cancer biomarkers (liquid biopsies) such as circulating tumor cells (CTCs) and their subpopulation circulating cancer stem cells (cCSCs) as an effective, feasible, repeatable, and non-invasive tool for tumor prognosis and treatment monitoring. In the current prototype study, we detected and enumerated the circulatory breast cancer stem cells (BCSCs) for the first time in the feline peripheral blood by using the flow cytometer analysis (FCA) against their specific cellular markers (CD44+/CD24- and CD133+). In addition, we calculated Area Under the Curve (AUC), like hood ratios and cutoff points by statistical analysis. The results revealed statistically significant differences (p <0.0001) between healthy and diseased animal groups with an excellent AUC value (0.902 for CD44+/CD24- cells and 0.990 for CD133+ population) that supported the use of FCA as a sensitive, specific, and rapid diagnostic and monitoring tool for mammary carcinoma. We also set up a cutoff value of diagnostic significance (>276 for CD44+/CD24- populations and >12 for CD133+ cells).
Keywords | Feline mammary tumor; Cancer Biomarkers; Tumor Stem Cells; CD133+; CD44+/CD24-, Flow cytometer.
Received | September 30, 2021; Accepted | November 05, 2021; Published | November 15, 2021
*Correspondence | Salah A. Selim, Haithem A. M. Farghali, Department of Microbiology, Faculty of Veterinary Medicine, Cairo University, 12211 Giza, Egypt; Department of Surgery, Anesthesiology and Radiology, Faculty of Veterinary Medicine, Cairo University,12211 Giza, Egypt; Email: [email protected];
Citation | Elsabagh RH, Farghali HAM, Emam IA, Ragab E, Nada AA, Selim SA (2021). Quantitative flow cytometry assessment of feline circulatory breast cancer stem cells. Adv. Anim. Vet. Sci. 9(12): 2201-2215.
DOI | http://dx.doi.org/10.17582/journal.aavs/2021/9.12.2201.2215
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2021 Farghali et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Spontaneous mammary tumors are one of the most common malignancies in cats. Almost 90% of the feline mammary carcinomas (FMC) are malignant, and they tend to metastasized and spread to the lungs and lymph nodes (Karabolovski et al., 2015). The WHO proposed female cats with naturally developed mammary tumors as an excellent model for studying breast carcinoma in human neoplasia. As feline shares many epidemiological, clinical, and morphological similarities with human breast cancer besides their shorter lifespan and the faster progression that provides a quicker gathering of data and trial accomplishment (De Las Mulas et al., 2000; Cannon, 2015).
The diagnosis, prognosis, and monitoring of therapeutic antitumor activity are mainly dependent on immunological examination or pathological features with histopathological investigations (Zappulli et al., 2015). The former techniques require repeatable tumor tissue biopsies for diagnosis, which are not feasible in all tumorigenic stages due to the critical sites of some tumors (Kazarian et al., 2017). As these procedures are time and effort-consuming, the early diagnosis of mammary tumors, employing serum/plasma biomarkers, seems crucial for the patient’s future (Kaszak et al., 2018). The serial analysis of liquid tumor biopsies is the best non-invasive, easily accessible method for breast cancer prediction, diagnosis, and prognosis (Duffy et al., 2018; Nicolini et al., 2018). Circulating Tumor Cells (CTCs) and their subpopulation Cancer Stem Cells (CSCs) were introduced by immense clinicians as an important prognostic marker for cancers, including Breast Cancer (BC). Being limited to the live tumor, unlike circulatory tumor DNA, gives CTCs prevalence over other blood biomarkers as they are illustrative for active tumor loci (Lopresti et al., 2019). Also, these cells can escape lysis by natural killer (NK) cells, survive in the bloodstream and resist apoptosis which support their metastatic abilities (Labelle et al., 2011; Freeman et al., 2015; Wu et al., 2017). These informations strongly supported that CTCs enumeration can be used as a prognostic index for patient overall survival rate. In humans, the cutoff value ≥ 5 cells per 7.5 mL of whole peripheral blood (PB) is a consolidated prognostic factor with strong metastatic potentials and poor clinical outcomes (Wu et al., 2017).
Since 2003, many studies have supported the role of breast cancer stem cells (BCSCs( or tumor-initiating cells (TICs) in metastasis mediation, tumor recurrence, and resistance of chemo and radiotherapies after Al-Hajj et al. used CD44+/CD24- surface markers combination for the first isolation of BCSCs (Al-Hajj et al., 2003; Sayed et al., 2016; Mansoori et al., 2017).
The cancer cell can acquire stemness during Epithelial-Mesenchymal Transition (EMT) by up-regulation of CD44v, ALDH, CD133, and epithelial cell adhesion molecule (EpCAM) and downregulation of CD24 and E-cadherin (E-cad), together with co-expression of epithelial markers (cytokeratins (CKs)) and mesenchymal markers (calponin (CALP)) (Jaggupilli and Elkord, 2012; Khoo et al., 2016; Granados-Soler et al., 2018). Breast neoplasia with high expression of CD44+/CD24-/low is associated with boosted invasion and metastasis and localized at the tumor’s invasive front (Baba and Câtoi, 2007; Liu et al., 2014). In comparison, those with high levels of CD133+ are associated with more aggressive and resistant tumor behavior and restricted deeply to intratumor hypoxic regions (Brugnoli et al., 2019).
Many previous studies that had been performed on companion animals have reported the identification and isolation of BCSCs. In felines and canines, TICs were identified and isolated by mammosphere formation, decrease ALDH1 enzyme activity, or cell surface markers (CD44, CD24, and CD133) either from spontaneous mammary carcinoma (Barbieri et al., 2012, 2015) or mammary carcinoma cell lines (Michishita et al., 2013; Pang et al., 2013).
Similar to humans, the isolated canine and feline BCSCs were invasive with a high potential for mammosphere formation, exhibited epithelial-mesenchymal transition (EMT) phenotype, and showed resistance to chemotherapy drugs and radiation (Pang et al., 2013; Barbieri et al., 2015). These proliferative and aggressive cancer cells showed a promising treatment response by nanoparticles (Gold nanoparticles (GNPs), silver nanoparticles (AgNPs)) in many previous studies in humans as well as felines (Ali et al., 2016; Sameen et al., 2020; Jabir et al., 2021; Jawad et al., 2021).
Because cBCSCs are a rare subpopulation in PB, their isolation and identification represent a real challenge (Yang, Imrali and Heeschen, 2015). As of now, there are two techniques to isolate potential BCSCs depending on their specific surface markers: either by fluorescence‐activated cell sorting (FACS) or magnetic‐activated cell sorting (MACS) (Akbarzadeh et al., 2019). The Flow Cytometer Analysis (FCA) is the most accurate, sensitive, and definitive laboratory tool for recognizing sporadic cells like BCSCs as it is capable of detecting one epithelial cell in up to 107 peripheral blood mononuclear cells (Hu et al., 2010).
In veterinary oncology, M. Michishita and coworkers (Michishita et al., 2013) were the first to use the FCA to recognize TICs in feline mammary carcinoma cell lines. Also, the diagnostic precision of FCA to discriminate neoplastic from non-neoplastic lymphoproliferative anarchies in cats was reported in a study by Martini and colleagues (Martini et al., 2018).
For the first time in cats, we proposed to detect and enumerate cBCSCs in feline peripheral blood using the FC analysis against their specific cellular markers: CD44+/CD24- combination and CD133+ individually. We aimed to evaluate the sensitivity and specificity of this technique for the early diagnosis of FMC using the receiver operating characteristic (ROC) curve and tried to set up a cutoff point of diagnostic value for each used phenotypic marker/ combination.
Materials and Methods
Animals groups
This study was carried out in compliance with the ARRIVE guidelines (du Sert, Ahluwalia, et al., 2020; du Sert, Hurst, et al., 2020), and Studies of diagnostic accuracy (STARD) guidelines (Bossuyt et al., 2015). All animals under investigation in the study were handled following the Association for Assessment and Accreditation of Laboratory Animal Care and Office of Laboratory Animal Welfare (AAALAC) guidelines under the direction of the Institutional Animal Care and Use Committee at Cairo University (CU-IACUC) that approved the protocol (code: CU II F 9 16) renewed by (Vet CU28/04/2021/266). Written informed consent was provided by the owners of the cats for recruitment in our research.
Between March 2019 and January 2021, 1ml whole blood samples were collected from each 26 client-owned female cats of different breeds that were admitted to the referral animal hospital of the Faculty of Veterinary Medicine at Cairo University.
We collected nine whole blood samples from 9 healthy queens that have been free of any disease (GA: healthy cats). The other 17 EDTA blood samples were collected from 17 female unspayed cats, diagnosed with mammary tumor (GB: diseased cats), as shown in (Table 1). All samples were stored at 4C° till analysis (within max 2-4hrs. post collection) (Diks et al., 2019).
Table 1: Descriptions of animal groups.
| Group name | Group description | Breeds | Total |
| GA | Healthy female cats with no mammary tumors or other pathological conditions |
3 Siamese 2 Mongrel 4 Persian |
9 |
| GB |
Female cats diagnosed positive with a mammary tumor (Note: 7 cats showed lung lesions) |
12 Persian 3 Siamese 2 Mongrel |
17 |
Then, all the diseased cats were introduced to a new treatment research study which will be published soon.
Diagnosis of mammary tumor in GB animals was performed by through physical examination, radiographic imagining for lung metastasis, and histopathological analysis to tumor biopsies by hematoxylin and eosin (H&E) and confirmed by Immunohistochemistry (IHC).
Physical and Radiographic Examination and Pathological studies
In the diseased cases (GB), we measured the dimensions of each tumor using calipers, and for detection of lung metastasis, we used the X-ray machine (Fischer, Berlin, Germany). The radiographic setting factors were as previously described (El-Rasikh et al., 2021). We followed the modified World Health Organization grading system to determine primary tumor size and the presence of lung metastasis (Cassali et al., 2019).
Tumor biopsies were collected from tumors> 3cm (T1& T2 categories) while tumors of the T3 category (> 3cm) (11 tumors in 7 diseased cases) have been surgically removed by our surgeons’ team in the Surgery Department, Faculty of Veterinary Medicine, Cairo University as previously mentioned (Farghali et al., 2017; Ali et al., 2019). We used Methadone (Comfortan® 10 mg/ml, 0.1 mg/Kg-0.3mg/Kg b.wt SC/IM; Dechra,UK ) every three to six hours post operative. The peri-operative measures were performed according to AAHA/AAFP pain management guidelines for dogs and cats (Epstein et al., 2015).
The pathological assessment of tumor samples was performed in the Pathology Department at Animal Health Research Institute (AHRI), Dokki, Giza. The formalin-fixed paraffin-embedded sections from tumor biopsies were processed routinely for H&E staining according to Suvarna. et al (S.Kim Suvarna , Christopher Layton, 1989) and Ali et al (Ali et al., 2018).
Further sections on positively charged coated slides were used for IHC (DAP- Peroxidase and Alkaline Phosphatase) technique using antibodies against (CD44 (CD44 Antibody IM7, Bio-Rad Laboratories) and CD133 (Recombinant Anti-CD133 antibody [EPR16508], Abcam, UK). These human antibodies were reported to have cross-reactivity with feline and canine tissues (Barbieri et al., 2012; Park, 2013; Granados-Soler et al., 2018). We used the BLASTP tool (BLASTP 2.12.0+) (Altschul et al., 2005) to assess the similarity between Human CD133 (O43490.1) and that of feline due to lake of evidence of cross reactivity antibodies. The query coverage was 96% with E-value (0.0) [S.1,2].
Proper negative controls were used according to The Histochemical Society’s standards (Hewitt et al., 2014). We could not assess the expression of CD24 because of the absence of antibody cross-reaction with the feline antigen. We implemented the IHC technique according to the kit’s manufacture instructions.
Flow Cytometer analysis
We analyzed whole PB samples to enumerate Circulatory Breast Cancer Stem Cells (cBCSCs) by flow cytometer (Coulter Epics XL, Beckman Coulter, CA) using Flow Jo software (BD) (Al-Salman et al., 2020). cBCSCs were detected against the following specific cell surface markers in each sample: CD133+ (CD133/1 (AC133) Antibody, anti-human, PE, REAlease, Miltenyi Biotec Germany) in a single test and CD44+/CD24- combination in another test (PE Mouse Anti-Human CD44v6 (2F10) BD Biosciences, USA and FITC Mouse Anti-Human CD24 (ML5), BD Biosciences, USA) according to their manufacture structure. We used CD44v6 antibody as it is nearly absent on most of leukocytes and many studies have approved it as a prognostic marker for BC (Mackay et al., 1994; Liu et al., 2016; Qiao et al., 2018).
The human antibodies used have already been reported to stain feline samples by FCA (Michishita et al., 2013; Pang et al., 2013). One tube served as a negative control (unstained cells) for each sample. Lysis of RBC and fixation of peripheral blood leukocytes after staining with specific antibodies were performed according to the Thermo Fisher Scientific flow cytometry protocol for staining cell surface antigens. Briefly, (1) 10 mL of diluted eBioscience™ 10X RBC Lysis Buffer (Multi-species) (Invitrogen, Thermofisher) were added to each PB sample (1 ml blood), mixed well and incubated at room temperature for 15 min, followed by centrifugation for 10 min. Then, the cell pellets were resuspended and washed three times by 2ml of eBioscience™ Flow Cytometry Staining Buffer (Invitrogen, Thermofisher) and resuspended in 300 µl of the same previous buffer. (2) For each 100 µl of the previous suspension, we added 20μL of eBioscience™ Fc Receptor Binding Inhibitor Polyclonal Ab, incubated for 15 min at room temperature, washed and resuspended in 300 µl of staining buffer. Then, 20 µl from each specific conjugated antibody (either CD133+ separately or CD44+/ CD24- combination) were added to each 100 µl of the previous suspension, vortexed gently, and incubated for 30 min at 4C° in the dark. About 2 mL of diluted eBioscience™ 10X RBC Lysis Buffer (Multi-species) (Invitrogen, Thermofisher) was added to the previous combination, incubated for 15 min in the dark, washed twice in the previously used staining buffer, then resuspended in 500 µl of this same reagent. (3)10-20 µl of eBioscience™ 7-AAD Viability Staining was added to the final cell suspension 15 min prior to sample acquisition on the cytometer.
cBCSCs were counted by recording all events in the whole suspension. BCSCs were identified as CD133+ or CD44+/ CD24- combination. The results are represented as mean ± standard deviation (SD), median, and range.
Statistical analysis
The data were checked for errors. Normality was checked using descriptive statistics, plots (histogram and box plot), and the Shapiro Wilk test. Comparison of the CD44+/CD24- and CD133+ absolute number of cells was implemented using the Mann Whitney U test. We used a binary logistic regression to define the cancer probability using the enumerated cellular markers. Area Under the Curve, sensitivity, specificity, and likelihood ratios, and cutoff points were calculated. Sensitivity values were plotted against complementary specificity values in the receiver operating characteristic (ROC) curve. The significance level was set at a p-value of 0.05 (Younus et al., 2019). For data analysis, we used MedCalc Statistical Software version 19.2.6 (MedCalc Software Ltd, Ostend, Belgium; https://www.medcalc.org;2020).
Results
Physical and radiographic examination
Physical examination of GB revealed about 39 tumors of different size categories and sites (Fig.1; A-C). Data considering the incidence of cats’ tumor numbers, sites, and sizes are shown in (Table 2). The radiographic examination diagnosed seven cats with lung metastasis (41.18%) among the diseased groups (Fig.1; D).
Table 2: Tumor characteristics of the diseased cases (GB).
| GB (n=39 tumors) | ||
| Tumor site: n (%) | Caudal abdomen | 11 (28.2%) |
| Caudal thoracic | 8 (20.5%) | |
| Cranial abdomen | 8 (20.5%) | |
| Cranial thoracic |
6 (15.4%) |
|
| Inguinal | 6 (15.4%) | |
| Side: n (%) | Left | 17 (43.6%) |
| Right | 22 (56.4%) | |
| Size category: n (%) | T1 (< 2cm) | 25 (64.1%) |
| T2 (2-3cm) | 3 (7.69%) | |
| T3 (> 3cm) |
11 (28.2%) |
|
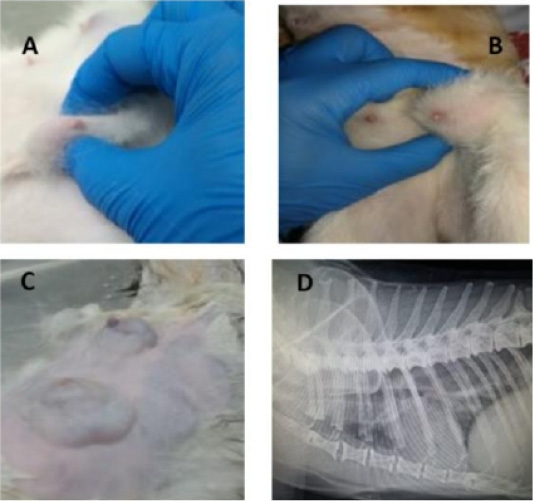
Figure 1: Feline physical and radiographic examination. Physical and radiographic examination of diseased animals (A-D). Queens with different tumor sizes (A-C), and Queen with lung metastasis (D).
Table 3: Statistical analysis of the (CD44+/CD24- ) and CD133+ Cells enumeration in the diseased and controls (GA &GB).
|
Diseased (n=17) |
Controls (n=9) |
P-value |
||
|
CD44+/CD24- cells |
Mean (SD) | 19003.06 (18714.61) | 1169.89 (90.67) | <0.0001* |
| Median (Min – Max) | 15765 (26 – 62043) | 161.00 (38 – 276) | ||
|
CD133+ cells |
Mean (SD) | 298.82 (640.205) | 5.00 (4.74) |
<0.0001* |
| Median (Min – Max) | 45.00 (13 – 2544) |
4.00 (0 – 14) |
||
*Statistically significant at p-value ≤0.05.
Table 4: Incidence of lung metastasis, Mean ± standard deviation (SD), median and range of (CD44+/CD24-) and CD133+ cells population in GB queens.
| Avarage | Lung Mets. n (%) | ||
| Yes | No | ||
| 7 (41.18%) | 10 (58.82%) | ||
|
(CD44+/CD24-) cells |
Mean (SD) | 24210.86 (15037.42) | 15357.60 (20881.40) |
| Median (Min-Max) | 21323 (5295 – 45784) | 4047 (26 – 62043) | |
|
P value |
0.143 | ||
|
CD133+ cells |
Mean (SD) | 176.71 (400.992) | 384.30 (775.715) |
| Median (Min-Max) | 28 (14 – 1086) | 89 (13 – 2544) | |
|
P value |
0.064 | ||
Table 5: Area Under the Curve for (CD44+/CD24-) and CD133+ cells “Diagnostic ability”.
| CD44+/CD24- | CD133 | ||
| AUC | Estimate | 0.902 | 0.990 |
| 95% CI | (0.721– 0.983) | (0.850 – 1.00) | |
|
P- value |
<0.0001* | <0.0001* | |
| Sensitivity | Estimate | 88.24 | 100 |
| 95% CI | 63.6 – 98.5 | 80.5 – 100 | |
| Specificity | Estimate | 100 | 88.89 |
| 95% CI | 66.4 – 100 | 51.8 – 99.7 | |
| LR+ | Estimate | -** | 9 |
| 95% CI | -** | 1.4 – 57.1 | |
| LR- | Estimate | 0.12 | 0 |
| 95% CI | 0.03 – 0.4 | - | |
| Youden index | Estimate | 0.88 | 0.89 |
| Cutoff point | Estimate | >276 |
>12 |
AUC: Area Under Curve, CI: Confidence Interval LR+: positive likelihood ratio, LR-: negative likelihood ratio. *Statistically significant at p-value ≤0.05. **Absence of positive LR is due to 100% specificity.
Pathological studies
All the examined tumor biopsies revealed features of mammary gland carcinoma (either ductal, tubular, or adenocarcinoma) when stained with H&E (Fig.2; A-C). In IHC, CD44 and CD133 demonstrated positive peroxidase reaction in all examined tissues either in the cell membrane of the lining epithelium (CD44) alone or with cytoplasm (CD133) (Fig.2; D&E).
Flowcytometry evaluation
The cells with the phenotypic expression of (CD44+/CD24- ) combination and CD133+ that correspond with breast cancer stem cells were detected and enumerated in each PB sample in this study (GA and GB) by using a flow cytometer (Fig.3). The count of (CD44+/CD24-) cells and CD133+ populations were significantly prevalent in cats diagnosed with mammary tumor (GB; n= 17) vs apparently healthy cats (GA; n=9) (p <0.0001). Mean ± standard
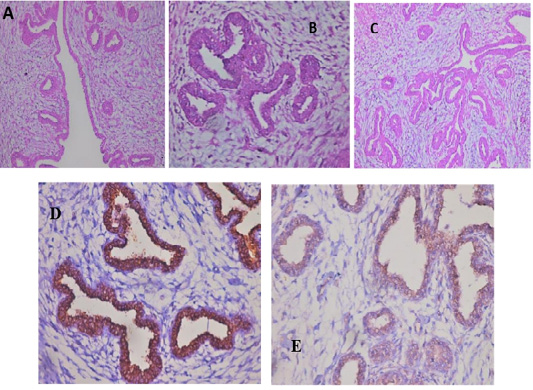
Figure 2: Feline mammary carcinoma. Representative pictures for H&E demonstrated hyperplasia of ductal epithelial with gland formation (A-C) (H&E: A, C X 200 & B X 400). Positive CD44 peroxidase reaction (brown color) in the cell membrane of lining epithelium (D) (CD44-peroxidase X 400). IHC showed a positive CD133 peroxidase reaction (brown color) in both cytoplasm and cell membrane of ductal lining epithelium (E) (CD133-peroxidase X 200).
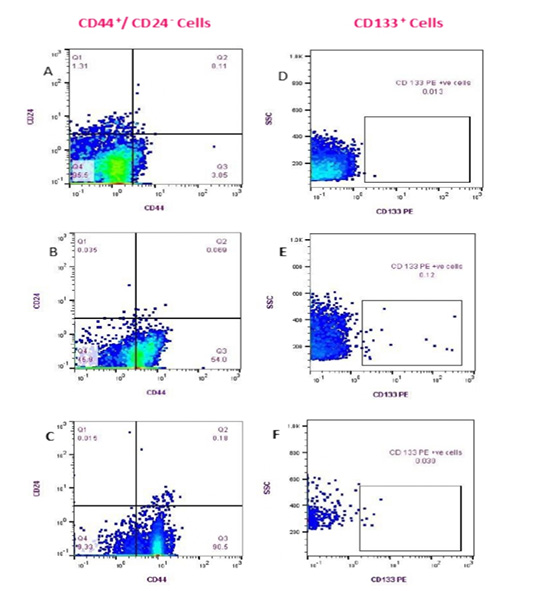
Figure 3: Flow cytometry detection of (CD44+/CD24- ) and CD133+ cell populations in Feline PB. (Fluorescence level of CD44 (PE) versus CD24 (FITC) (A-C)); Healthy cat (A), a diseased cat without lung metastasis (B) and diseased cat with lung metastasis (C). The data are representative of one cat in each animal group (GA, GB). Fluorescence level of CD133 (PE) versus cellular complexity (SS: SS) (D-F); Healthy cat (D), Diseased cat without lung metastasis (E), and diseased cat with lung metastasis (F).
deviation (SD), median, and range of CD44+/CD24- cells and CD133+ phenotypes of control and diseased groups are shown in (Table 3). Queens that diagnosed with lung metastasis (n:7) showed remarkable increase in the (CD44+/CD24-) cell populations over the rest of the GB (n:10). On the contrary, the CD133+ cells count was relatively higher in female cats without metastasis than those showed lung metastasis (Table 4).
Evaluation of diagnostic potency of (CD44+/CD24−) and CD133+ cells enumeration in feline mammary tumors
The receiver operating characteristic (ROC) analysis (Table 5) revealed an excellent AUC value for both (CD44+/CD24− ) and CD133+ phenotypes (excellent Ref. ratio between 0.9 and 1) with 100% sensitivity of CD133+ cells and 88.24% of (CD44+/CD24− ) cells (Fig. 4). The optimal cutoff points of (>276) for (CD44+/CD24− ) cells and (>12) for CD133+ cells generate the high Youden index of 0.88and 0.89, respectively (Table 5).
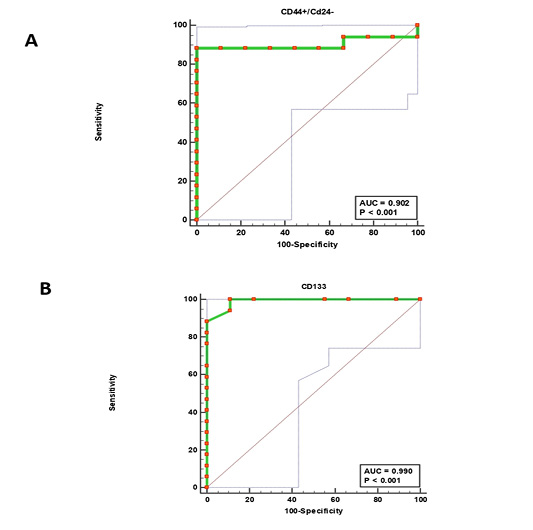
Figure 4: Receiver operating characteristic (ROC) curves. (CD44+/CD24-) cells (A) and CD133+ cells (B).
Table 6: Binary logistic regression.
| OR | 95% CI |
P-value |
|
|
(CD44+/CD24-) cells |
1.004 | (0.997 – 1.010) | 0.297 |
|
CD133+ cells |
2.240 | (0.251 – 19.956) | 0.470 |
Dependent variable: Ref category is having cancer. OR: odds ratio. CI: Confidence Interval.
The Odds Ratios (OR) (Table 6) of the binary logistic regression analysis evaluated the correlation between cancer occurrence and cBCSCs counts with (1.004) OR for (CD44+/CD24−) population and (2.240) OR for CD133+ cells. However, these results are not significant (P<0.05).
Discussion
CTCs and their subpopulation BCSCs are characteristic of،،Active،، tumor points (that potentially initiate metastasis) (Lopresti et al., 2019). Consequently, accurate enumeration of BCSCs with FCA depending on their specific cellular markers (CD44+/CD24-) and /or CD133+) is important.
Herein, we document the first time the flow cytometry quantitative analysis of BCSCs in the PB of cats diagnosed with the mammary tumor. The results showed that BCSCs could be detected and counted in the PB of feline depending on their definite cellular markers ((CD44+/CD24-) and CD133+); FCA is a sensitive, specific, and rapid diagnostic tool for mammary carcinoma and we also established a cutoff value of diagnostic significance for both (CD44+/CD24-) and CD133+ populations.
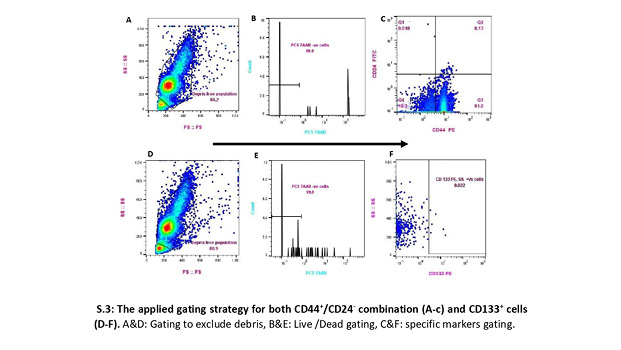
To overcome the paucity of BCSCs, we imitated the flow cytometry analysis technique previously reported (Al-Hajj et al., 2003; Martini et al., 2020). This strategy included all events in the whole suspension, verified by a back-gating method based on cellular morphologic properties (forward scatter-height [FSC-H] vs. side scatter-height [SSC-H]), excluding dead cells, and with the proper acquisition, we adjusted the speed according to the number of events to enhance detection sensitivity [S.3].
In the current prototype, (CD44+/CD24−) and CD133+ cell populations were detected in PB of all animals under investigation with a statistically significant difference between diseased group (n=17) and healthy queens (n=9) (p <0.0001).
These flow cytometry outcomes were utterly matched with our results of the initial clinical and IHC diagnosis. The same diseased positive groups gave statistically significant higher values in the flow cytometry analysis. Our findings are similar in humans as many studies validate the BCSC expression as a diagnostic and prognostic marker for aggressive breast cancers (Matsuoka and Yashiro, 2015; Sayed et al., 2016; Mansoori et al., 2017; Lee et al., 2019; Elbaiomy et al., 2020).
Previous studies have proposed flow cytometry as the best laboratory tool for detecting rare cell populations like BCSCs in blood as it can identify cells at frequencies as low as 0.0001% (Goodale et al., 2009; Watanabe et al., 2014). Our ROC curve analysis strongly supported this conclusion by a sensitivity of 88.24% for (CD44+/CD24−) cell population and 100% for CD133+ cell population. While specificity was 100% and 88.89%, respectively. Furthermore, the ROC curve gave an excellent AUC value of 0.902 (95% CI: 0.721– 0.983) and 0.990 (95% CI: 0.850 – 1.00) for (CD44+/CD24−) and CD133+ expressions, respectively (Table 5). These presented findings confirm the former conclusion about the diagnostic accuracy of FCA to distinguish tumorigenic from non-tumorigenic lymphoproliferative disorders in cats (Martini et al., 2018).
For the first time in cats, we set up a diagnostic significant cutoff value for the measurements of cBCSCs in 1 mL of feline peripheral blood, which is >276 for (CD44+/CD24-) cells and >12 for CD133+ cells that generated a Youden index of 0.88 and 0.89, respectively. In humans, no estimated diagnostic cut-off value for cBCSCs was established yet. However, prognostic cutoff points of 2 CTCs/4ml blood in one study and 3CTCs/1ml blood in another have been reported (Lopresti et al., 2019; Jin et al., 2020).
The same is also noticed among canine investigations; there is no clear estimated diagnostic cutoff point for cBCSCs. However, a value of zero CTCs in the healthy negative control dogs was reported in a study by L. Marconato. et al (Marconato et al., 2019).
The stress of handling and speed during the venipuncture and the procedure site might affect hematology results by shifting up the RBC and elevating the WBC count, which might explain the relatively high values of the established cutoff points in the feline. So, it is recommended to restrict sampling to one person with standardized procedures (O’Brien et al., 1998). Upon this conclusion, we cannot exclude such an effect on CTCs and their subpopulation (cBCSCs). Also, on luminal epithelial cells of the feline malignant mammary tumor, the upregulation of P-Cadherin and loss of expression or abnormal function of E-cadherin increase tumor cell proliferation, motility, invasiveness, high infiltrative growth, and the presence of neoplastic emboli that consequently increase CTCs and cCSCs populations in queens with mammary tumor (Figueira et al., 2014).
We also applied Binary Logistic Regression analysis for the first time in queens to assess the strength of association between CD44+/CD24- or CD133+ counts and having breast cancer as independent factors. The odds of having a mammary tumor were 1.004 times as likely to have one CD44+/CD24-cell and 2.240 times as likely to have one CD133+ cell. Previous studies have measured the strength of the association between BCSCs and disease prognosis in humans (Elbaiomy et al., 2020), and canines (Marconato et al., 2019). Nevertheless, this association between cBCSCs and having mammary tumors is not statistically significant (P<0.05) (Table 6), which may be related to the relatively low number of animals used in this study.
Previous studies have proposed the role of Epithelial-Mesenchymal Transition (EMT) during the starting of the metastasis process (Yang et al., 2015). EMT enables carcinoma cells to gain migratory and invasive abilities together with obtaining stemness characteristics. This transition helps tumor cells escape from the primary tumor lesions, intravasate into the circulation, and disseminate at distant secondary tumor sites (Mani et al., 2008; Yilmaz and Christofori, 2009). Tumor epithelial cells that undergo EMT show upregulation of CD44 down-regulation of CD24 and other stem cell markers (Mani et al., 2008). These events could explain the remarkable increase of (CD44+/CD24-) cell population in the diseased cats with lung metastasis more than the rest of the diseased group. Thus, our results came compatible with previous studies which confirmed that the (CD44+/CD24- /low) cell population is associated with boosted invasion and metastasis (Baba and Câtoi, 2007), and so we are suggesting that (CD44+/CD24-/low) cell enumeration could be used as a prognostic tool for metastasis initiation and treatment monitoring in the feline.
Several former articles demonstrated the ability of neoplastic cells to adapt to the state of low oxygen accessibility (hypoxia). Almost half of breast cancer lesions, enfold many hypoxic regions varying in amount and size (Bhandari et al., 2019). These hypoxic regions are reported to be associated with clinically aggressive tumor behavior (Walsh et al., 2014). The expression of CD133 was informed to be induced by low oxygen availability. Therefore, an increase in CD133expression can be considered an aggressive tumor evolution marker with prognostic and predictive values (Brugnoli et al., 2019), and might explain why, in our investigation, the CD133+ cells count was relatively higher in female cats without metastasis than those with metastasis.
Conclusion
In conclusion, during this study, we assessed the diagnostic significance of the flow cytometer enumeration of cBCSCs for the first time in the feline PB. The (ROC) analysis revealed excellent AUC rates (0.902 and 0.990) for both (CD44+/CD24− ) and CD133+ phenotypes respectively. Also, we established a cutoff point of diagnostic values for both (CD44+/CD24- /low) and CD133+ cell populations (>276 and >12), respectively. In addition, our team used the Odds Ratio to measure the strength of the association between (CD44+/CD24- /low) and CD133+ cells detection and mammary tumor occurrence that was (1.004) OR for (CD44+/CD24−) population and (2.240) OR for CD133+ cells.
Since our investigation was based on a relatively low number of subjects, we recommend applying the mentioned FCA diagnostic technique to a large-scale animal population and investigating any correlation between tumor size, sites, numbers, and circulatory BCSCs count. However, we noticed a nonsignificant correlation between lung metastasis and upregulation of (CD44+/CD24- /low) cells in queen’s circulation, further separate studies about the relation of FC enumeration of the cBCSCs and feline mammary tumor metastasis are recommended.
In addition, we endorse including cases with benign mammary conditions such as benign tumors or hyperplasia to evaluate the ability of FCA to discriminate between benign and malignant diseases. We also suggest running a comparative analysis between the number of cCSC and their number in the solid tumor besides applying the previous diagnostic regime as a model in human breast oncology studies.
One of this study’s limitations; is that we can’t run a single flow panel with both CD44+, CD24-, and CD133+ as the available CD44 and the CD133 antibodies were of the same fluorophore (PE).
Acknowledgments
We thank Prof. Dr. Naglaa M. Alkalamawy (Pathology Department at Animal Health Research Institute (AHRI), Egypt) for examination and imaging the pathological slides. We thank Assistant lecturer; Hams Abdelrahman (Dental Public Health Department at Alexandria University, Egypt) for performance of all statistical analysis. We thank Christopher P Cook and Aymen Khalifa (Department of Dermatology, University of California, San Francisco, San Francisco CA, USA) for their generous contribution in editing of flow cytometry analysis.
Conflict of Interest
No potential competing of interest was reported by authors.
authors contribution
Data curation, Rasha Elsabagh and Ibrahim Emam; Funding acquisition, Rasha Elsabagh and Haitham Fargali; Investigation, Rasha Elsabagh and Ibrahim Emam; Methodology, Haitham Fargali; Resources, Rasha Elsabagh, Haitham Fargali and Ibrahim Emam; Supervision, Haitham Fargali, Eman Ragab and Abdelfattah Nada; Validation, Rasha Elsabagh and Haitham Fargali; Visualization, Rasha Elsabagh; Writing – original draft, Rasha Elsabagh; Writing – review & editing, Haitham Fargali. Salah A. Selim; Conceptualization, Methodology, Writing - Review & Editing, Supervision, Project administration.
References
S1:
RID: F9K3BM4K01R
Job Title:sp|O43490|
Program: BLASTP
Query: RecName: Full=Prominin-1; AltName: Full=Antigen AC133; AltName: Full=Prominin-like protein 1; AltName: CD_antigen=CD133; Flags: Precursor ID: O43490.1(amino acid) Length: 865
Database: RefSeq protein Felis catus Refseq Protein
Sequences producing significant alignments:
Scientific Common Max Total Query E Per. Acc.
Description Name Name Taxid Score Score cover Value Ident Len Accession
prominin-1 isoform X3 [Felis catus] Felis catus domestic cat 9685 1179 1179 96% 0.0 66.71 849 XP_023109054.1
prominin-1 isoform X1 [Felis catus] Felis catus domestic cat 9685 1171 1171 96% 0.0 66.23 855 XP_019685284.2
prominin-1 isoform X4 [Felis catus] Felis catus domestic cat 9685 1159 1159 96% 0.0 65.99 840 XP_019685292.2
prominin-1 isoform X2 [Felis catus] Felis catus domestic cat 9685 1157 1157 94% 0.0 66.75 849 XP_019685287.2
Alignments:
>prominin-1 isoform X3 [Felis catus]
Sequence ID: XP_023109054.1 Length: 849
Range 1: 15 to 844
Score:1179 bits(3050), Expect:0.0,
Method:Compositional matrix adjust.,
Identities:559/838(67%), Positives:693/838(82%), Gaps:9/838(1%)
Query 14 CGNSFSGGQPSSTDAPKAWNYELPATNYETQDSHKAGPIGILFELVHIFLYVVQPRDFPE 73
Sbjct 15 ...TLCL.SR...EGTEVLEL.........K..Y......G..QI.....R....N.... 74
Query 74 DTLRKFLQKAYESKIDYDKPETVILGLKIVYYEAGIILCCVLGLLFIILMPLVGYFFCMC 133
Sbjct 75 .I...I...KFDLST..E...N.V.T...I...I.V.I.A......V.......CC.GL. 134
Query 134 RCCNKCGGEMHQRQKENGPFLRKCFAISLLVICIIISIGIFYGFVANHQVRTRIKRSRKL 193
Sbjct 135 ...............K..........V.....S.F.....I.......HL.AQ.EKT... 194
Query 194 ADSNFKDLRTLLNETPEQIKYILAQYNTTKDKAFTDLNSINSVLGGGILDRLRPNIIPVL 253
Sbjct 195 .....R.......GA.AE.S...S..T...E...S..DNVK.L.....HEQ...KV..A. 254
Query 254 DEIKSMATAIKETKEALENMNSTLKSLHQQSTQLSSSLTSVKTSLRSSLNDPLCLVHPSS 313
Sbjct 255 .D..A..E.........L.V.N...E.KKS.A..NT..SD..RD.EQ.....M.SAP.VA 314
Query 314 ETCNSIRLSLSQLNSNPELRQLPPVDAELDNVNNVLRTDLDGLVQQGYQSLNDIPDRVQR 373
Sbjct 315 T...N..T.....DD.TNMD...SL.KPI.KI.DI.Q.N.SS......K.F....EM.EN 374
Query 374 QTTTVVAGIKRVLNSIGSDIDNVTQRLPIQDILSAFSVYVNNTESYIHRNLPTLEEYDSY 433
Sbjct 375 ...DIISDV.ST........ESIGEQI....Q..N.MG.I.D..T..R............ 434
Query 434 WWLGGLVICSLLTLIVIFYYLGLLCGVCGYDRHATPTTRGCVSNTGGVFLMVGVGLSFLF 493
Sbjct 435 R......V.C....V...........T.....N....R.........I.....A.V...I 494
Query 494 CWILMIIVVLTFVFGANVEKLICEPYTSKELFRVLDTPYLLNEDWEYYLSGKLFNKSKMK 553
Sbjct 495 .....T.......V.......L....QNRK..Q..........N.KH....MV...PDIN 554
Query 554 LTFEQVYSDCKKNRGTYGTLHLQNSFNISEHLNINEHTGSISSELESLKVNL-NIFLLGA 612
Sbjct 555 ...........E.K.I....K.E..Y........Q..A.N..NDFQNM...ID..V..DE 614
Query 613 AGRKNLQDFAACGIDRMNYDSYLAQTGKSPAGVNLLSFAYDLEAKANSLPPGNLRNSLKR 672
Sbjct 615 ......M..SFS...AI..NI...ELS.A.TKG......D...V...H..H.S.KQ...N 674
Query 673 DAQTIKTIHQQRVLPIEQSLSTLYQSVKILQRTGNGLLERVTRILASLDFAQNFITNNTS 732
Sbjct 675 N....R...RSE.I.L...M.SV...I.E..QKSS..GMK..NT.S...S..D.L.TRI. 734
Query 733 SVIIEETKKYGRTIIGYFEHYLQWIEFSISEKVASCKPVATALDTAVDVFLCSYIIDPLN 792
Sbjct 735 ...V..SQ...N..V....R....VKI..T..I.A.........S.............M. 794
Query 793 LFWFGIGKATVFLLPALIFAVKLAKYYRRMDSEDVYDDVETIPMKNMENGNNGYHKDH 850
Sbjct 795 .........................H..........E.--------.....I.F.RH. 844
>prominin-1 isoform X1 [Felis catus]
Sequence ID: XP_019685284.2 Length: 855
>prominin-1 isoform X1 [Felis catus]
Sequence ID: XP_019685285.2 Length: 855 >prominin-1 isoform X1 [Felis catus]
Sequence ID: XP_019685286.2 Length: 855
Range 1: 15 to 850
Score:1171 bits(3029), Expect:0.0,
Method:Compositional matrix adjust.,
Identities:559/844(66%), Positives:693/844(82%), Gaps:15/844(1%)
Query 14 CGNSFSGGQPSSTDAPKAWNYELPATNYETQDSHKAGPIGILFELVHIFLYVVQPRDFPE 73
Sbjct 15 ...TLCL.SR...EGTEVLEL.........K..Y......G..QI.....R....N.... 74
Query 74 DTLRKFLQKAYESKIDYDKPETVILGLKIVYYEAGIILCCVLGLLFIILMPLVGYFFCMC 133
Sbjct 75 .I...I...KFDLST..E...N.V.T...I...I.V.I.A......V.......CC.GL. 134
Query 134 RCCNKCGGEMHQRQKENGPFLRKCFAISLLVICIIISIGIFYGFVANHQVRTRIKRSRKL 193
Sbjct 135 ...............K..........V.....S.F.....I.......HL.AQ.EKT... 194
Query 194 ADSNFKDLRTLLNETPEQIKYILAQYNTTKDKAFTDLNSINSVLGGGILDRLRPNIIPVL 253
Sbjct 195 .....R.......GA.AE.S...S..T...E...S..DNVK.L.....HEQ...KV..A. 254
Query 254 DEIKSMATAIKETKEALENMNSTLKSLHQQSTQLSSSLTSVKTSLRSSLNDPLCLVHPSS 313
Sbjct 255 .D..A..E.........L.V.N...E.KKS.A..NT..SD..RD.EQ.....M.SAP.VA 314
Query 314 ETCNSIRLSLSQLNSNPELRQLPPVDAELDNVNNVLRTDLDGLVQQGYQSLNDIPDRVQR 373
Sbjct 315 T...N..T.....DD.TNMD...SL.KPI.KI.DI.Q.N.SS......K.F....EM.EN 374
Query 374 QTTTVVAGIKRVLNSIGSDIDNVTQRLPIQDILSAFSVYVNNTESYIHRNLPTLEEYDSY 433
Sbjct 375 ...DIISDV.ST........ESIGEQI....Q..N.MG.I.D..T..R............ 434
Query 434 WWLGGLVICSLLTLIVIFYYLGLLCGVCGYDRHATPTTRGCVSNTGGVFLMVGVGLSFLF 493
Sbjct 435 R......V.C....V...........T.....N....R.........I.....A.V...I 494
Query 494 CWILMIIVVLTFVFGANVEKLICEPYTSKELFRVLDTPYLLNEDWEYYLSGKLFNKSKMK 553
Sbjct 495 .....T.......V.......L....QNRK..Q..........N.KH....MV...PDIN 554
Query 554 LTFEQVYSDCKKNRGTYGTLHLQNSFNISEHLNINEHTGSISSELESLKVNL-NIFLLGA 612
Sbjct 555 ...........E.K.I....K.E..Y........Q..A.N..NDFQNM...ID..V..DE 614
Query 613 AGRKNLQDFAACGIDRMNYDSYLAQTGKSPAGVNLLSFAYDLEAKANSLPPGNLRNSLKR 672
Sbjct 615 ......M..SFS...AI..NI...ELS.A.TKG......D...V...H..H.S.KQ...N 674
Query 673 DAQTIKTIHQQRVLPIEQSL------STLYQSVKILQRTGNGLLERVTRILASLDFAQNF 726
Sbjct 675 N....R...RSE.I.L...MKYGKAR.SV...I.E..QKSS..GMK..NT.S...S..D. 734
Query 727 ITNNTSSVIIEETKKYGRTIIGYFEHYLQWIEFSISEKVASCKPVATALDTAVDVFLCSY 786
Sbjct 735 L.TRI....V..SQ...N..V....R....VKI..T..I.A.........S......... 794
Query 787 IIDPLNLFWFGIGKATVFLLPALIFAVKLAKYYRRMDSEDVYDDVETIPMKNMENGNNGY 846
Sbjct 795 ....M..........................H..........E.--------.....I.F 846
Query 847 HKDH 850
Sbjct 847 .RH. 850
>prominin-1 isoform X4 [Felis catus]
Sequence ID: XP_019685292.2 Length: 840
Range 1: 15 to 835
Score:1159 bits(2998), Expect:0.0,
Method:Compositional matrix adjust.,
Identities:553/838(66%), Positives:686/838(81%), Gaps:18/838(2%)
Query 14 CGNSFSGGQPSSTDAPKAWNYELPATNYETQDSHKAGPIGILFELVHIFLYVVQPRDFPE 73
Sbjct 15 ...TLCL.SR...EGTEVLEL.........K..Y......G..QI.....R....N.... 74
Query 74 DTLRKFLQKAYESKIDYDKPETVILGLKIVYYEAGIILCCVLGLLFIILMPLVGYFFCMC 133
Sbjct 75 .I...I...KFDLST..E.---------.I...I.V.I.A......V.......CC.GL. 125
Query 134 RCCNKCGGEMHQRQKENGPFLRKCFAISLLVICIIISIGIFYGFVANHQVRTRIKRSRKL 193
Sbjct 126 ...............K..........V.....S.F.....I.......HL.AQ.EKT... 185
Query 194 ADSNFKDLRTLLNETPEQIKYILAQYNTTKDKAFTDLNSINSVLGGGILDRLRPNIIPVL 253
Sbjct 186 .....R.......GA.AE.S...S..T...E...S..DNVK.L.....HEQ...KV..A. 245
Query 254 DEIKSMATAIKETKEALENMNSTLKSLHQQSTQLSSSLTSVKTSLRSSLNDPLCLVHPSS 313
Sbjct 246 .D..A..E.........L.V.N...E.KKS.A..NT..SD..RD.EQ.....M.SAP.VA 305
Query 314 ETCNSIRLSLSQLNSNPELRQLPPVDAELDNVNNVLRTDLDGLVQQGYQSLNDIPDRVQR 373
Sbjct 306 T...N..T.....DD.TNMD...SL.KPI.KI.DI.Q.N.SS......K.F....EM.EN 365
Query 374 QTTTVVAGIKRVLNSIGSDIDNVTQRLPIQDILSAFSVYVNNTESYIHRNLPTLEEYDSY 433
Sbjct 366 ...DIISDV.ST........ESIGEQI....Q..N.MG.I.D..T..R............ 425
Query 434 WWLGGLVICSLLTLIVIFYYLGLLCGVCGYDRHATPTTRGCVSNTGGVFLMVGVGLSFLF 493
Sbjct 426 R......V.C....V...........T.....N....R.........I.....A.V...I 485
Query 494 CWILMIIVVLTFVFGANVEKLICEPYTSKELFRVLDTPYLLNEDWEYYLSGKLFNKSKMK 553
Sbjct 486 .....T.......V.......L....QNRK..Q..........N.KH....MV...PDIN 545
Query 554 LTFEQVYSDCKKNRGTYGTLHLQNSFNISEHLNINEHTGSISSELESLKVNL-NIFLLGA 612
Sbjct 546 ...........E.K.I....K.E..Y........Q..A.N..NDFQNM...ID..V..DE 605
Query 613 AGRKNLQDFAACGIDRMNYDSYLAQTGKSPAGVNLLSFAYDLEAKANSLPPGNLRNSLKR 672
Sbjct 606 ......M..SFS...AI..NI...ELS.A.TKG......D...V...H..H.S.KQ...N 665
Query 673 DAQTIKTIHQQRVLPIEQSLSTLYQSVKILQRTGNGLLERVTRILASLDFAQNFITNNTS 732
Sbjct 666 N....R...RSE.I.L...M.SV...I.E..QKSS..GMK..NT.S...S..D.L.TRI. 725
Query 733 SVIIEETKKYGRTIIGYFEHYLQWIEFSISEKVASCKPVATALDTAVDVFLCSYIIDPLN 792
Sbjct 726 ...V..SQ...N..V....R....VKI..T..I.A.........S.............M. 785
Query 793 LFWFGIGKATVFLLPALIFAVKLAKYYRRMDSEDVYDDVETIPMKNMENGNNGYHKDH 850
Sbjct 786 .........................H..........E.--------.....I.F.RH. 835
>prominin-1 isoform X2 [Felis catus]
Sequence ID: XP_019685287.2 Length: 849
Range 1: 15 to 838
Score:1157 bits(2992), Expect:0.0,
Method:Compositional matrix adjust.,
Identities:550/824(67%), Positives:683/824(82%), Gaps:7/824(0%)
Query 14 CGNSFSGGQPSSTDAPKAWNYELPATNYETQDSHKAGPIGILFELVHIFLYVVQPRDFPE 73
Sbjct 15 ...TLCL.SR...EGTEVLEL.........K..Y......G..QI.....R....N.... 74
Query 74 DTLRKFLQKAYESKIDYDKPETVILGLKIVYYEAGIILCCVLGLLFIILMPLVGYFFCMC 133
Sbjct 75 .I...I...KFDLST..E...N.V.T...I...I.V.I.A......V.......CC.GL. 134
Query 134 RCCNKCGGEMHQRQKENGPFLRKCFAISLLVICIIISIGIFYGFVANHQVRTRIKRSRKL 193
Sbjct 135 ...............K..........V.....S.F.....I.......HL.AQ.EKT... 194
Query 194 ADSNFKDLRTLLNETPEQIKYILAQYNTTKDKAFTDLNSINSVLGGGILDRLRPNIIPVL 253
Sbjct 195 .....R.......GA.AE.S...S..T...E...S..DNVK.L.....HEQ...KV..A. 254
Query 254 DEIKSMATAIKETKEALENMNSTLKSLHQQSTQLSSSLTSVKTSLRSSLNDPLCLVHPSS 313
Sbjct 255 .D..A..E.........L.V.N...E.KKS.A..NT..SD..RD.EQ.....M.SAP.VA 314
Query 314 ETCNSIRLSLSQLNSNPELRQLPPVDAELDNVNNVLRTDLDGLVQQGYQSLNDIPDRVQR 373
Sbjct 315 T...N..T.....DD.TNMD...SL.KPI.KI.DI.Q.N.SS......K.F....EM.EN 374
Query 374 QTTTVVAGIKRVLNSIGSDIDNVTQRLPIQDILSAFSVYVNNTESYIHRNLPTLEEYDSY 433
Sbjct 375 ...DIISDV.ST........ESIGEQI....Q..N.MG.I.D..T..R............ 434
Query 434 WWLGGLVICSLLTLIVIFYYLGLLCGVCGYDRHATPTTRGCVSNTGGVFLMVGVGLSFLF 493
Sbjct 435 R......V.C....V...........T.....N....R.........I.....A.V...I 494
Query 494 CWILMIIVVLTFVFGANVEKLICEPYTSKELFRVLDTPYLLNEDWEYYLSGKLFNKSKMK 553
Sbjct 495 .....T.......V.......L....QNRK..Q..........N.KH....MV...PDIN 554
Query 554 LTFEQVYSDCKKNRGTYGTLHLQNSFNISEHLNINEHTGSISSELESLKVNL-NIFLLGA 612
Sbjct 555 ...........E.K.I....K.E..Y........Q..A.N..NDFQNM...ID..V..DE 614
Query 613 AGRKNLQDFAACGIDRMNYDSYLAQTGKSPAGVNLLSFAYDLEAKANSLPPGNLRNSLKR 672
Sbjct 615 ......M..SFS...AI..NI...ELS.A.TKG......D...V...H..H.S.KQ...N 674
Query 673 DAQTIKTIHQQRVLPIEQSL------STLYQSVKILQRTGNGLLERVTRILASLDFAQNF 726
Sbjct 675 N....R...RSE.I.L...MKYGKAR.SV...I.E..QKSS..GMK..NT.S...S..D. 734
Query 727 ITNNTSSVIIEETKKYGRTIIGYFEHYLQWIEFSISEKVASCKPVATALDTAVDVFLCSY 786
Sbjct 735 L.TRI....V..SQ...N..V....R....VKI..T..I.A.........S......... 794
Query 787 IIDPLNLFWFGIGKATVFLLPALIFAVKLAKYYRRMDSEDVYDD 830
Sbjct 795 ....M..........................H..........EE 838