Advances in Animal and Veterinary Sciences
Research Article
New Epizootic of Lumpy Skin Disease in Assiut- Egypt: Molecular Identification and Characterization
Khaled A. S. El-Khabaz1*, Eman A. M. Shosha2, Aml M. Abdel-Ra’ouf3
1Department of Animal Medicine (Infectious Diseases), Faculty of Veterinary Medicine, Assiut University, Egypt; 2Department of Microbiology and Immunology, Faculty of Veterinary Medicine, New Valley University, Egypt; 3Department of Animal Medicine (Infectious Diseases), Faculty of Veterinary Medicine, Aswan University, Egypt.
Abstract | An outbreak of lumpy skin disease (LSD) was recorded during the period of 2018 in the study area. Tissue samples from skin nodules were surgically collected from diseased animals (N=23) for molecular identification of the causative virus by conventional polymerase chain reaction (PCR) using specific primers designated from G protein coupled chemokine receptor (GPCR) gene. All the skin samples were found to be positive for lumpy skin disease virus (LSDV) as they gave the expected amplicon size at 554 bp. Sequencing analysis of the obtained amplified segment disclosed that the present outbreak virus has a very high identity percentage >99-100% with virulent LSDV either that’s isolated from Egypt or different foreign countries, while the percent of identity were 95.62% with the vaccinal strains of LSDV. Multiple alignment and Phylogenetic analysis of the obtained sequence manifesting that members of capripoxviruses could be segregated in different groups and the present outbreak strain grouped within the LSDV virulent strains cluster.
Keywords | Lumpy skin disease, capripoxvirus, GPCR gene, phylogenetic analysis, PCR
Received | December 07, 2020; Accepted | December 17, 2020; Published | January 15, 2021
*Correspondence | Khaled AS El-Khabaz, Department of Animal Medicine (Infectious Diseases), Faculty of Veterinary Medicine, Assiut University, Egypt; Email: [email protected]
Citation | El-Khabaz KAS, Shosha EAM, Abdel-Raouf AM (2021). New epizootic of lumpy skin disease in assiut- egypt: molecular identification and characterization. Adv. Anim. Vet. Sci. 9(3): 446-452.
DOI | http://dx.doi.org/10.17582/journal.aavs/2021/9.3.446.452
ISSN (Online) | 2307-8316; ISSN (Print) | 2309-3331
Copyright © 2021 El-Khabaz et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Lumpy skin disease (LSD) is a highly important insect-borne cattle disease caused by a virus belonging to family poxviridae, genus capripox. The genus capripox include lumpy skin disease virus (LSDV), sheeppox virus (SPPV), and goatpox virus (GTPV), which are genetically related and host-specific (Le Goff et al., 2005).
Clinically LSD characterize by rise of body temperature, anorexia, appearance of intradermal skin nodules all over the body, loss of condition and enlargement of superficial lymph nodes (Ochwo et.al., 2020). There was a variation in the intensity of clinical signs according to pathogenicity of causative virus, breed, age, immunity, and production state of host, and vector spreading (OIE, 2018).
Diagnosis of LSD is usually based upon the pathognomic clinical signs and different laboratory techniques such as, virus isolation and identification, several PCR techniques and serological diagnosis. Serological diagnosis is time-devouring and cannot differentiate between different members of capripoxviruses (El-Bagoury et al., 2009).
The first outbreak of LSD in Egypt was in the year 1988 after the importation of cattle from Somalia (House et al., 1990) after that, several epidemics were recorded in different years (El-Kholy et al., 2008; Raof et al., 2010; Sharawi and Abd El-Rahim, 2011; El-Khabaz, 2014; Tamam et al., 2018; Rouby et al., 2019; Hodhod, 2020).
Previously, the disease was a threat to Africa only, but now it is well known that it has been spread to Asia (Rashid et. al., 2017; Kasem et.al., 2018) and Europe (Tasioudi et.al., 2016; Toplak et al., 2017; Kononov et al., 2019).
GPCR are transmembrane proteins play a great role in the transmission of signals of various stimuli such as light, odorants, nucleotide and proteins from extracellular to intracellualar (Kostenis, 2004). Capripoxviruses homologue of GPCR responsible for cell proliferation in addition to suppression of the antiviral effect elicited by the host (Cao et al., 1995; Tulman et al., 2002). By phylogenetic analysis of GPCR gene, members of genus capripoxvirus can be clearly segregated into different obvious sets (Le Goff et al., 2009).
This study aims to molecularly identify lumpy skin disease virus, which has been circulating in Assiut governorate during the year 2018, by partial amplification of GPCR gene. Also sequencing and phylogenetic analysis of the amplified gene sequence with a subsequent comparison between the obtained LSDV sequences and these available in GenBank.
Materials and methods
Study area
Assiut governorate which is located in Upper Egypt, characterized by hot and relatively humid weather during summer and slightly cold with rear rains during winter.
Ethical approval
The examination and sampling were done without causing any exertion or hurt to the animals.
Animals
A total of 23 diseased native and mixed breed cattle were used in this study.
Clinical examination
The clinical examination of all diseased animals under study was carried out according to Jackson and Cockcroft (2002).
Samples
Including all skin layers (epidermis, dermis, and subcutis), skin nodules biopsies (n=23) were collected aseptically from different 23 diseased cattle of different ages and from different localities of Assiut governorate. All skin nodules samples were kept at -20 ᵒC in a deep freezer till used.
DNA extraction
DNA was extracted from skin nodules by using G-spin DNA extraction Kit (Intron Cat. no. P53077) according to the manufacturer’s instructions. The extracted DNA was then stored at –20 °C for subsequent PCR assay.
Primer
The primers were designated from G-Protein Coupled Chemokine Receptor (GPCR) gene according to (Tamam et al., 2018) with the following sequences:
Forward primer 5′- AGT ACA GTT AGT AGC GCA ACC -3′ and reverse primer: 5′- GGG TGA ACT ACA GCT AGG TAT-3′, this is to amplify 554 bp.
Polymerase chain reaction
It was performed according to kit manual instructions using the thermal cycler (Biorad gradient temperature minicycler). The parameters were: initial denaturation at 95°C for 10 minutes, 40 cycles (denaturation at 95 °C for 30 seconds, annealing at 50°C for 30 seconds, extension at 72°C for 1 minute); and a final extension phase run at 72°C for 10 min.
Electrophoreses of PCR products were done in a 2% agarose gel. The agarose gel was prepared by dissolving 1 gram agarose in 50 ml 1X TAE buffer and heated using microwave till becoming semisolid. 2μl ethidium bromide (0.5mg/ml) (INtRON Biotechnology, Korea) was added to each 50ml agarose solution. 10µL of the PCR products as well as the DNA ladder were loaded on the gel after mixing with loading dye, and visualized by UV transilluminator and photographed (BIORAD, USA).
Sequencing and phylogenetic analysis
By using QIA quick gel extraction kit (Qiagen, Germany), one of the positive samples by conventional PCR was purified and sequenced in both directions using the same primer. Sequencing was done by 3500 XL Genetic Analyzer in the Genome Research Unit- Animal Health Research Institute (AHRI- Giza, Egypt).
Accession number
The gained partial GPCR gene sequence was submitted to the Genbank under Accession No. MT793047.
Database search
The obtained nucleotide sequence of partial GPCR gene of LSDV was searched in the nucleotide collection standard databases (National Center for Biotechnology Information, NCBI) using megaBLAST optimized for highly similar sequences option.
Multiple alignment and phylogenetic analysis
Multiple alignments were created with clustal W integrated into Molecular Evolutionary Genetics Analysis (MEGA) version X software. The phylogenetic tree was constructed by the use of maximum-likelihood method based on Tamura-Nei model in Molecular Evolutionary Genetics Analysis (MEGA) version X software (Kumar et al., 2018) with Bootstraping confidence calculated at 1000 replicates.
Results
Clinical features
All the examined animals in this study were showing the characteristic clinical features of lumpy skin disease with variation in severity ranged from mild to severe. The commonly noticed clinical signs were fever, an apparition of skin nodules with variable distribution patterns with necrosis, and sloughing of many of them leaving deep ulcers (Figure 1), loss of condition, and anorexia. In addition to the previous signs, respiratory distress, tarry like diarrhea, edema in forelimbs and/or brisket, and corneal opacity were observed in some of the examined animals in different degrees.
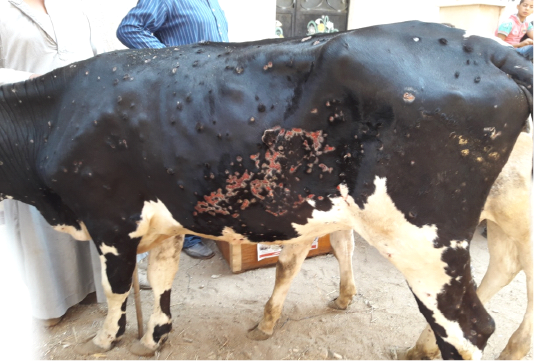
Figure 1: Multiple intradermal skin nodules with sloughing of many of them leaving deep ulcers due to infection with LSDV
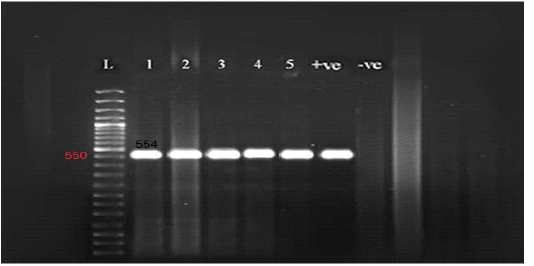
Figure 2: Gel electrophoresis of amplified partial GPCR gene Lane L (50 bp Ladder marker), lanes 1-5 (positive samples), lane +ve (control positive), and lane –ve (control negative).
Molecular identification
All the examined skin nodules samples (n=23) from diseased animals were found to be positive for LSD virus when tested by conventional PCR using a specific primer for the GPCR gene. The partial GPCR gene of LSDV was successfully amplified as the anticipated amplicon size 554 bp was clearly recorded in all skin samples from all diseased animals (Figure 2).
Sequencing and phylogenetic analysis
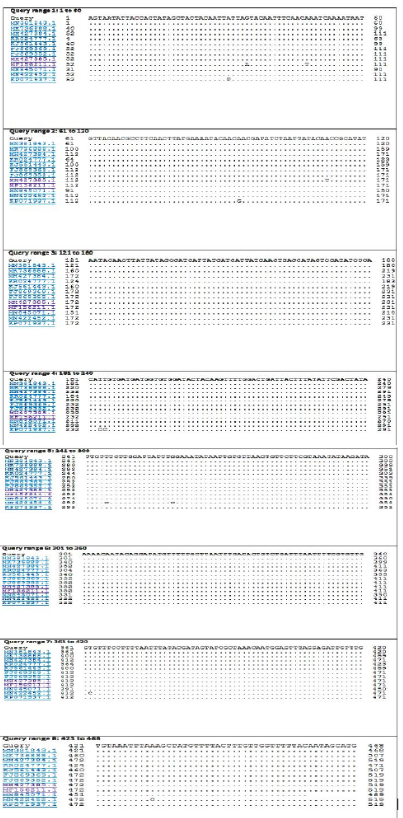
The BLASTn search analysis showing that the present outbreak sequence of partial GPCR gene has a very high percentage of identity >99-100% with all virulent LSDV isolated from different outbreaks in Egypt (KJ561443.1,
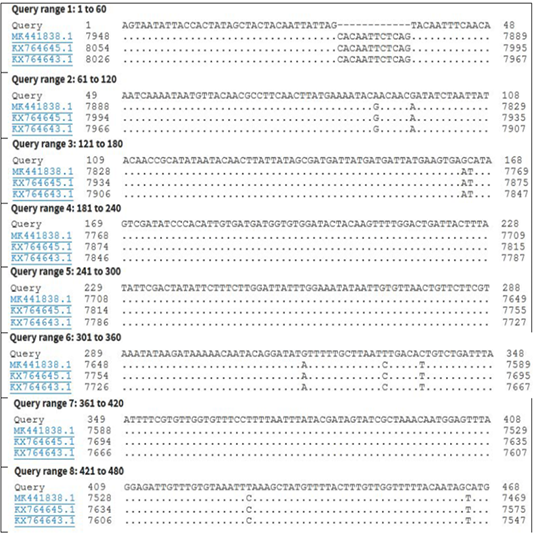
Figure 4: Nucleotide sequence alignment identity of partial GPCR gene of the present LSDV outbreak isolate with different LSDV vaccinal isolates.
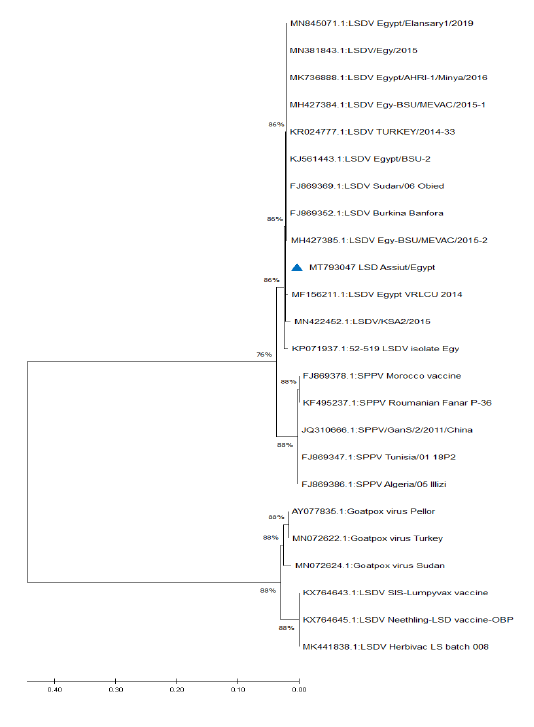
Figure 5: Phylogenetic analysis of different capripoxviruses based upon the nucleotide sequences of GPCR gene.
KP071937.1, MF156211.1, MH427384.1, MK736888.1, MH427385.1, MN845071.1, and MN381843.1), and in some different countries such as Turkey (KR024777.1), Sudan (FJ869369.1), KSA (MN422452.1), and Burkina Faso (FJ869352.1) (Figure 3). This percent of identity lowered to be 95.62% with LSDV vaccinal strains (Figure 4). The generated phylogenetic tree has shown four genetically clear clusters lineages (LSDV field isolates, SPPV, GTPV, and LSDV vaccinal strains) (Figure 5), and our epidemic strain (labeled with blue triangle) is found in the LSDV field isolates group.
Discussion
In Egypt, where LSD is endemic, reliance placed on vaccination using the sheep pox vaccine. Obviously, there were some faults as many epidemics appear at different time intervals. During the summer of 2018, a new epizootic of LSD has been noticed, affecting cattle of different ages and of both sexes, in different localities of the Assiut governorate. The present work directed toward molecular identification and characterization of the causative agent of such an outbreak.
In the present study, all the examined animals were found exhibiting the characteristic clinical signs of lumpy skin disease such as a sharp increase in body temperature above the normal level, firm nodules covering a large area of the body surface with necrosis and tearing out some of them, decreased appetite and milk production. Further clinical abnormalities such as corneal opacity, lymphadenopathy, edematous swelling in forelegs, and tarry-like diarrhea were observed in some of the diseased animals. Previously, similar clinical signs were recorded by Tuppurainen et al. (2005), Gari et al. (2010), Tasioudi et al. (2016), Tassew et.al. (2018), Rouby et al. (2019). For confirmation of these clinical suspicious, a conventional PCR (using specific primer) was employed to amplify 554 bp region of GPCR gene LSDV. The results of PCR confirmed that the causative agent of such an outbreak is LSDV. Different studies reported that LSDV was responsible for several epidemics in different Egyptian governorates along the previous few years (El-Bagoury et al., 2009; El-Khabaz, 2014; Abdallah et al., 2018; El-kady et al., 2018; Hodhod et al., 2020). This outbreak was recorded in the study area during summer months where both the temperature and relative humidity are high with an increase of insect population. Humidity and temperature greatly increase the population of insect vector (Ayelet et. al., 2014; Zeynalova et.al., 2016; Kasem et.al., 2018).
For its speed and accuracy, PCR is an ideal test for the diagnosis of LSD in infected animals as it efficiently detects LSDV DNA in skin samples. PCR is a very credible test for the diagnosis of LSD for its velocity, sensitivity, and specificity (El-Kholy et al., 2008; Tamam et al., 2018). In the present study, all skin samples collected from diseased animals gave positive results by PCR. LSDV is present in higher concentrations and for long period in the skin so skin samples are more superior than blood or organs for the detection of viral DNA of LSDV (Zeynalova et al., 2016).
Due to the continuous importation of live cattle, especially from African countries, the molecular investigation of the origin of LSDV is very important from the epidemiological point of view. The genetic characters of the recognized LSDV, from examined animals in Assiut governorate, were evaluated by interpreting the phylogenetic tree and sequence identity. The obtained sequence and all the Egyptian field isolates were found in the same cluster, indicating that the same strain circulating in different governorates of Egypt, also the stable nature of LSDV. LSDV genome characterized by high conserved nature despite its circulation in different geographical regions (Sameea Yousefi et al., 2018; Adedeji et al., 2019).
Database search and blast analysis declare very high sequence conformity >99-100% of this outbreak LSDV sequence and the Egyptian virulent isolates deposited in Genbank. The phylogenetic analysis of the attained sequence of partial GPCR gene was capable to assort capripoxviruses into 4 distinct clusters (LSDV field isolates, SPPV, GTPV, and LSDV vaccinal strains); this is greatly supported with 1000 bootstrap reiterates confidence. Similar findings were recorded by Adedeji et al. (2019) and Ochwo et al. (2020). The field LSDV lineage, including the present outbreak strain, is more closely linked to SPPV, while the vaccinal isolates were very tightly associated with GTPV. LSDV is more correlated to SPPV than to GTPV (Beard et al., 2010; El-Kenawy and El-Tholoth, 2010; Rashid et al., 2017), while Le Goff et al. (2009) reported that LSDV was closely connected with GTPV rather than SPPV. Genetic studies proved that members of genus capripoxvirus had been arisen from one common ancestor during their evolution journey (Tulman et al., 2002; Le Goff et al., 2009).
In the current study the phylogenetic analysis based on the GPCR gene showing that all of the field isolates of LSDV were located in separate cluster away from the vaccinal strains, this seems to be helpful in differentiation between the field isolates from the vaccinal ones. Complete GPCR gene sequence analysis may be needed to confirm or deny this hypothesis.
Conclusion
The first step of controlling infectious diseases depends mainly upon the prompt and precise diagnosis, so it seems that GPCR based PCR is a very useful technique in the diagnosis of LSD in susceptible animals. The LSDV causing this outbreak was widely linked to different virulent strains detected in other Egyptian governorates. Sequencing of GPCR gene isn’t only useful in differentiation between different members of capripoxviruses but also the distinction between filed and vaccinal strains of LSDV.
Conflict of interest
All the authors declare that they have no conflict of interest.
Acknowledgment
Nothing to declare.
authors contribution
KASE and EAMS designed the study and edited the manuscript. KASE and AMA collected the samples. All authors shared in the lab work. KASE performed the interpretation of the results. The final version of the manuscript was read and approved by all authors.
References